Complete Mitochondrial Genome of Myra affinis (Decapoda:Brachyura: Leucosiidae) and Its Phylogenetic Implications for Brachyura
ZHANG Ying, MENG Lei, MIAO Zengliang, WEI Liming, LIU Bingjian,LIU Liqin, GONG Li, and Lü Zhenming, *
Complete Mitochondrial Genome of(Decapoda:Brachyura: Leucosiidae) and Its Phylogenetic Implications for Brachyura
ZHANG Ying1), MENG Lei2), MIAO Zengliang1), WEI Liming1), LIU Bingjian1),LIU Liqin1), GONG Li1), and Lü Zhenming1), *
1),,,,316022,2),,316022,
Knowledge of complete mitochondrial genomes (mitogenomes) can help understand the molecular evolution and phylogenetic relationships of various species. To date, the phylogenetic status of Leucosiidae within Brachyura remains unresolved because of the limited number of mitogenomes available. In the present study, the complete mitogenome ofwas sequenced using next-generation sequencing to supplement the limited mitogenome information of Leucosiidae.The 15349bp-long mitogenome includes 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a putative control region. The overall base composition is 34.5% A, 36.1% T, 11.5% G, and 17.9% C, with a high AT bias (70.6%). All 13 PCGs start with the standard ATN codon and stop with the TAN codon or incomplete T. Except for, all other tRNAs have a typical cloverleaf-like secondary structure. Phylogenetic analyses based on the maximum likelihood and Bayesian inference methods are employed to generate identical phylogenetic topologies, thereby supporting the sister relationship between Leucosiidae and Matutidae for the first time. The monophyly of Eubrachyura is well established, and its sister relationship with Raninoida is strongly supported.The results of this work will not only help achieve a better understanding of the characteristics of themitogenome and the phylogenetic position of Leucosiidae, but also provide relevant information for further studies on the phylogeny of Brachyura.
leucosiid crab; mitogenome; phylogenetic analysis; Brachyura
1 Introduction
The typical metazoan mitochondrial genome (mitoge- nome) is a closed-circular molecule of 14–20kb. It general-ly contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (and), and an AT-rich region (also called the control region, CR) (Boore, 1999). Compared with the nuclear genome, the mi- togenome is more widely employed in population genetics,comparative genomics, and phylogenetic studies (Sanchez., 2016; Tan., 2018; Ruan., 2020). The main advantages of the use of mitogenomes in molecular sys- tematics are as follows: 1) the small genome size and high genome copy numbers of mitogenomes reduce the diffi- culties of PCR amplification; 2) the relatively high evolu- tionary rate and low level of recombination of these ge- nomes are suitable for conducting evolutionary and phy- logenetic analyses; and 3) the different evolutionary rates of mitochondrial genes render them efficient materials in the phylogenetic analyses at different levels (., family, genus, or population). The advent and maturation of next- generation sequencing (NGS) technologies in recent years have markedly improved the ability to obtain and analyze mitogenomic data, which can further expand the popula- rity of mitogenomic research (Kumar., 2020; Ruan., 2020; Wang., 2020a).
The infraorder Brachyura contains approximately 7250 known species inhabiting marine, freshwater, and terres- trial habitats (Zhang, 2011; Chen., 2018; Ma., 2019).Brachyura, as the oldest crab, originated in the Ju- rassic period (Schweitzer and Feldmann, 2010; Davie., 2015), and a group of its members with extremely diverse morphology and ecology was finally formed after massive radiative evolution. However, this same diversity has also caused the identification of species of this order to be re- markably challenging, and their real phylogenetic relation- ships remain unknown (Rocha., 2018; Tan., 2018).The classification of Brachyura has a long history and has undergone several changes.Brachyura was initially seg- mented into three groups, namely, Podotremata, Heterotre-mata, and Thoracotremata (Spears., 1992).Subse-quently, the infraorder was proposed to be divided into Dro-miacea and Eubrachyura (including Thoracotremata, Rani- noida, and Heterotremata) (Martin and Davis, 2001). How- ever, the latest classification scheme divides Brachyura intoEubrachyura, Dromiacea, Raninoida, and Cyclodorippoida(Ahyong., 2007; Tsang., 2014).Although the phy- logenetic relationship within Brachyura is incompletely un-derstood, the current classification system has been recog- nized by most scholars.Leucosiidae, a diversified group belonging to Eubrachyura, is usually referred to as ‘peb- ble crab’ on account of its unusual carapaces, which vary from pyriform to oval and subcircular; this feature is quite different from that of traditional crabs (Naderloo, 2017). The habitats of this group are also diversified, and indivi- dual species may be found in coastal sand bottoms, gravel, muds, and nearshore watersin Asian countries, such as China, Japan, and Korea (De Melo, 1996).The phyloge- netic relationships among Leucosiidae and even the evo- lutionary status of this family have not been well resolved. Furthermore, only one complete mitogenome sequence of this family ()is available (Park., 2017). This lack of relevant data mainly hampers phylogenetic studies of Leucosiidae and even Brachyura.
The genusLeach, 1817, has been a source of sys- tematic and nomenclatural confusion (Galil, 2001).Bell, 1855,is a species mainly distributed in the Indo-West Pacifica, including most areas of the East Chi- na Sea and the South China Sea (Naderloo, 2017).Thus far, most studies of this species have focused on its mor- phology (Tan, 1996; Naderloo, 2017), and related mole- cular research is unavailable. In the present study, we first sequence and annotate the complete mitogenome ofto supplement the number of Leucosiidae mitoge- nomes currently available. The molecular features of this mitogenome, including mitogenome organization, codon usage, and tRNA secondary structures, are then described in detail. Next, the phylogeny of Brachyura is reconstruct- ed according to the nucleotide sequences of 13 PCGs from83 Brachyuran species and two Anomuran species (as out- groups). The data presented in this study will help achi- eve a better understanding of the characteristics of themitogenome, determine the phylogenetic position of Leucosiidae, and shed light on the evolutionary relation- ships among Brachyura.
2 Materials and Methods
2.1 Ethics Statement
The crab specimen used in this study was collected fromZhoushan Province, China (29?45?32??N, 121?45?30??E). The species was not involved in the endangered list of the International Union for Conservation of Nature (https:// www.iucnredlist.org/). Specimen collection and mainte- nance were performed in strict accordance with the rec- ommendations of Animal Care Quality Assurance in Chi- na. All experimental protocols were approved by the In- stitutional Ethics Committee of Zhejiang Ocean Univer- sity.
2.2 DNA Extraction, Mitogenome Sequencing,and Assembly
An SQ Tissue DNA Kit (OMEGA, USA) was used to extract the total genomic DNA from the muscle tissue of a single sample following the manufacturer’s instructions. The mitogenome ofwas sequenced by NGS of paired reads measuring 150bp in length (Illumina HisSeq 4000; Shanghai Origingene Bio-pharmTechnology Co., Ltd., China).Adapters and low-quality bases were removed us- ing Cutadapt v1.16 (Martin, 2011) with the following pa- rameters: ?q 20 ?m 20.Trimmed reads shorter than 50bp were discarded. Quality control of the raw and trimmed reads was performed using FastQC v0.11.5 (http://www. bioinformatics.babraham.ac.uk/projects/fastqc/). The filter- ed clean data were assembled and mapped to a complete mitogenome sequence using NOVOPlasty v2.7.2 (Dierck- xsens., 2017).
2.3 Mitogenome Annotation and Sequence Analyses
The complete mitogenome was manually annotated us- ing Sequin software (version 15.10, http://www.ncbi.nlm. nih.gov/Sequin/), and PCGs were determined by their openreading frames following the guidelines of the inverte- brate mtDNA translation table. The boundaries of rRNA and tRNA genes were determined using NCBI-BLAST (http://blast.ncbi.nlm.nih.gov) and tRNAscan-SE 1.21 (Lowe and Chan, 2016), respectively, and compared with the re- lated species. Transfer RNA (tRNA) genes were manually plotted according to the secondary structure predicted by tRNAscan-SE 1.21 (Lowe and Chan, 2016) and MITOS Web Server (Bernt., 2013) with invertebrate mito- chondrial genetic codes. The CR was determined by the locations of adjacent genes. The mitogenome map was drawn by CGView Server V 1.0 (Stothard and Wishart, 2005), and the base composition and relative synonymous codon usage (RSCU) were obtained using MEGA X (Ku- mar., 2018). The following formulas were used to calculate strand asymmetries (Perna and Kocher, 1995):

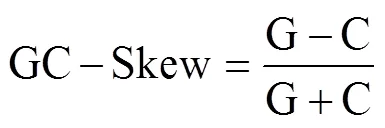
2.4 Phylogenetic Analysis
A total of 84 complete mitogenome sequences down- loaded from GenBank database (https://www.ncbi.nlm.nih. gov/genbank) and a newly determined sequence () were used to reconstruct the phylogenetic relation- ships among Brachyura (Table 1). In addition, two species from Anomura were used as outgroups (Table 1). The nu- cleotide sequences of 13 PCGs for each species were ex- tracted from the relevant GenBank files using PhyloSuite (Zhang., 2020a), and the MAFFT program (Katoh., 2002) integrated with PhyloSuite was executed to align multiple sequences into normal-alignment mode. Gblocks was used to identify and remove ambiguously aligned sequences using default settings (Talavera and Ca- stresana, 2007).The sequences were then concatenated and used to generate input files (phylip and nexus format) for phylogenetic analyses. Maximum likelihood (ML) and Ba- yesian inference (BI) were employed for phylogenetic ana- lyses. The best-fitting model was selected using Model- Finder (Kalyaanamoorthy., 2017) on the basis of the Bayesian Information Criterion. GTR+F+G7 and GTR+F+I+G4 were selected as the best-fitting models for ML and BI analyses, respectively. ML analysis was carried out in IQ-TREE (Nguyen., 2015) using an ML+rapid bootstrap (BS) algorithm with 1000 replicates.BI analy- sis was performed in MrBayes 3.2.6 (Ronquist., 2012) with default parameters and 3×106Metropolis-coupled Markov Chain Monte Carlo generations.Trees were sampled every 1000 generations with a burn-in rate of 25%. The average standard deviation of split frequencies below 0.01 was considered to reach convergence.
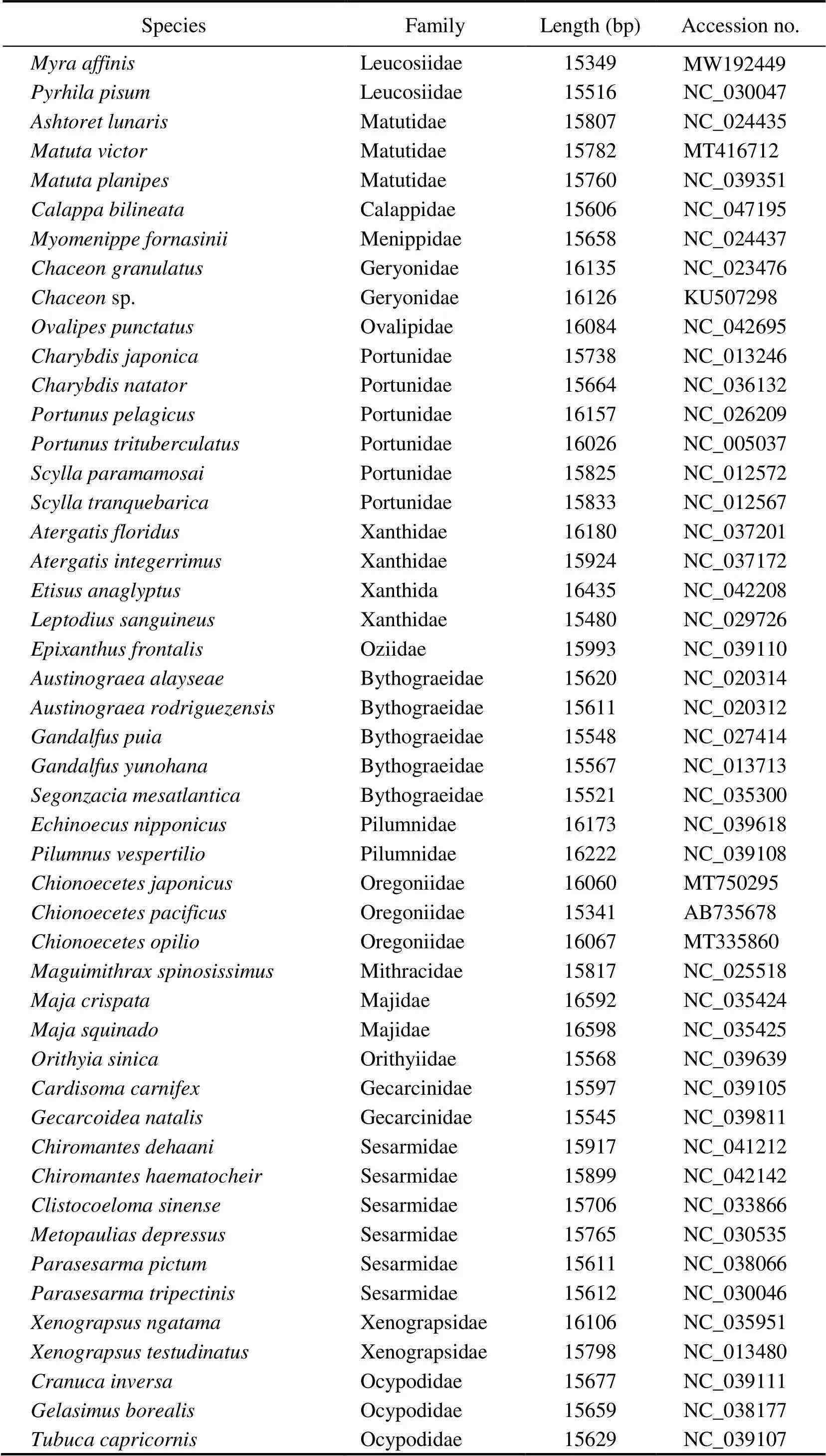
Table 1 List of 83 Brachyuran species and two outgroups used in this paper
()
()

SpeciesFamilyLength (bp)Accession no. Tubuca politaOcypodidae15672NC_039106 Ocypode ceratophthalmusOcypodidae15564NC_025324 Ocypode cordimanusOcypodidae15604NC_029725 Dotilla wichmanniDotillidae15600NC_038180 Ilyoplax deschampsiDotillidae15460NC_020040 Grapsus tenuicrustatusGrapsidae15858NC_029724 Pachygrapsus crassipesGrapsidae15652NC_021754 Pachygrapsus marmoratusGrapsidae15406NC_039109 Metopograpsus frontalisGrapsidae15587NC_042152 Metopograpsus quadridentatusGrapsidae15517NC_038178 Eriocheir hepuensisVarunidae16335NC_011598 Eriocheir sinensisVarunidae16354NC_006992 Eriocheir japonicaVarunidae16352NC_011597 Neoeriocheir leptognathusVarunidae16143NC_041211 Hemigrapsus penicillatusVarunidae16486NC_038202 Hemigrapsus sanguineusVarunidae16275NC_035307 Macrophthalmus darwinensisMacrophthalmidae16348MF457408 Macrophthalmus japonicusMacrophthalmidae16170NC_030048 Macrophthalmus pacificusMacrophthalmidae17226NC_046039 Mictyris longicarpusMictyridae15548NC_025325 Geothelphusa dehaaniPotamidae18197NC_007379 Geothelphusa sp.Potamidae18052MG674171 Huananpotamon lichuanensePotamidae15380NC_031406 Longpotamon yangtsekiensePotamidae17885NC_036946 Longpotamon kenliensePotamidae18499NC_044413 Potamiscus motuoensisPotamidae17971KY285013 Somanniathelphusa boyangensisGecarcinucidae17032NC_032044 Lyreidus brevifronsRaninidae16112NC_026721 Umalia orientalisRaninidae15466NC_026688 Ranina raninaRaninidae15557NC_023474 Homola orientalisHomolidae16084KT182071 Homologenus malayensisHomolidae15793NC_026080 Moloha majoraHomolidae15903NC_029361 Latreillia validaLatreilliidae15097MK204361 Dynomene pilumnoidesDynomenidae16475KT182070 Pagurus nigrofasciaPaguridae15392NC_042412 Pagurus gracilipesPaguridae16051LC222534
3 Results and Discussion
3.1 Mitogenome Organization
The complete mitochondrial genome offorms a closed-circular molecule measuring 15349bp in size (Gen- Bank accession no. MW192449). This mitogenome con- sists of 13 PCGs, 2 rRNAs, 22 tRNAs, and one putative CR (Fig.1, Table 2).Except for four PCGs (.,,,, and), eight tRNAs (.,-,,,Leu,,,, and), and two rRNAs, which aredistributed on the light (L-) strand, the other mitogenes aredistributed on the heavy (H-) strand (Table 2).Overall, the genes in themitogenome are closely arranged with interval (10 intergenic spacers totaling 146bp) and overlapping (11 overlaps totaling 26bp) phenomena (Ta- ble 2).Three typical overlaps occur between protein- coding genes (., 1bp betweenand, 7bp be- tweenand, 1bp betweenand; Ta- ble 2), and these overlaps are commonly identified in other crabs (Lu., 2020; Zhang., 2020b). As is the case with other Brachyuran mitogenomes (Wang., 2020a; Wang., 2020b; Zhang., 2020b), themi- togenome exhibits a high AT bias (70.6%).The AT- and GC-skews of the mitogenome are negative at ?0.022 and ?0.217, respectively (Table 3), thereby indicating that Ts and Cs are more abundant than As and Gs.
3.2 PCGs and Codon Usage
The mitogenome ofcontains 13 PCGs in the order typically found in most Brachyuran species and con- sists of seven NADH dehydrogenases (–and), three cytochrome c oxidases (-), two AT- Pases (and8), and one cytochrome b ().These 13 PCGs have a total length of 11092bp and encode 3687 amino acids. All of the 13 PCGs are initiated by the canonical start codon ATN. The majority of the 13 PCGs terminate with TAA or TAG, while four other PCGs (.,,,,and) use a single T as a stop codon(Table 2). Incomplete stop codons are common in meta- zoan mitogenomes and may be recoveredpost-trans- criptional polyadenylation (Ojala., 1981). The AT- and GC-skews of the 13 PCGs are similarly negative, at ?0.018 and ?0.004, respectively (Table 3). This finding de- monstrates that Ts and Cs are more abundant than As and Gs in the entire protein-coding gene sequence. Four PCGs (.,,,, and) have positive GC-skew values, thereby indicating that they are encoded by the L-strand, whereas the other nine PCGs exhibit negative values, which indicates they are encoded by the H-strand (Table 3).
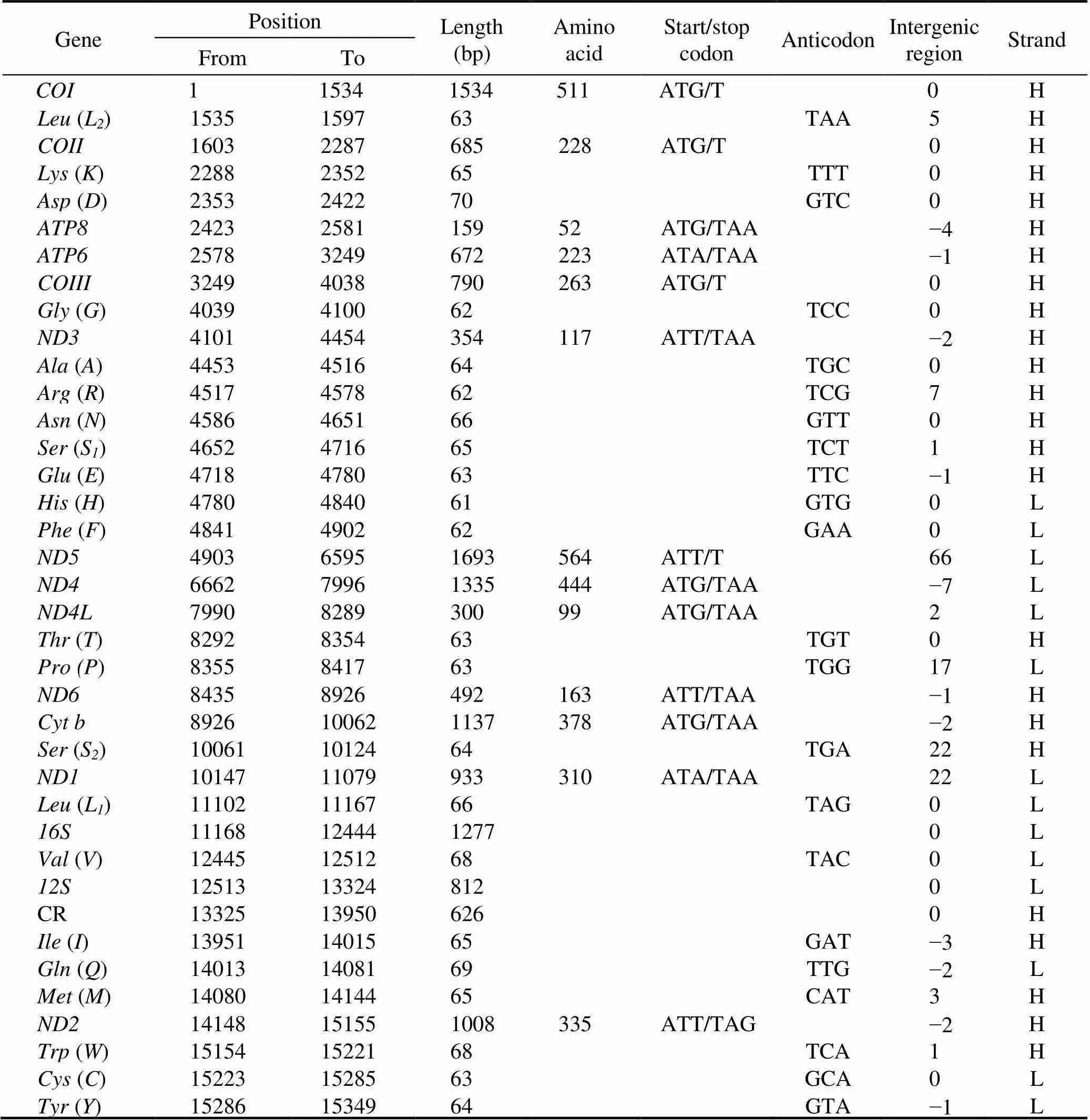
Table 2 Features of the M. affinis mitogenome
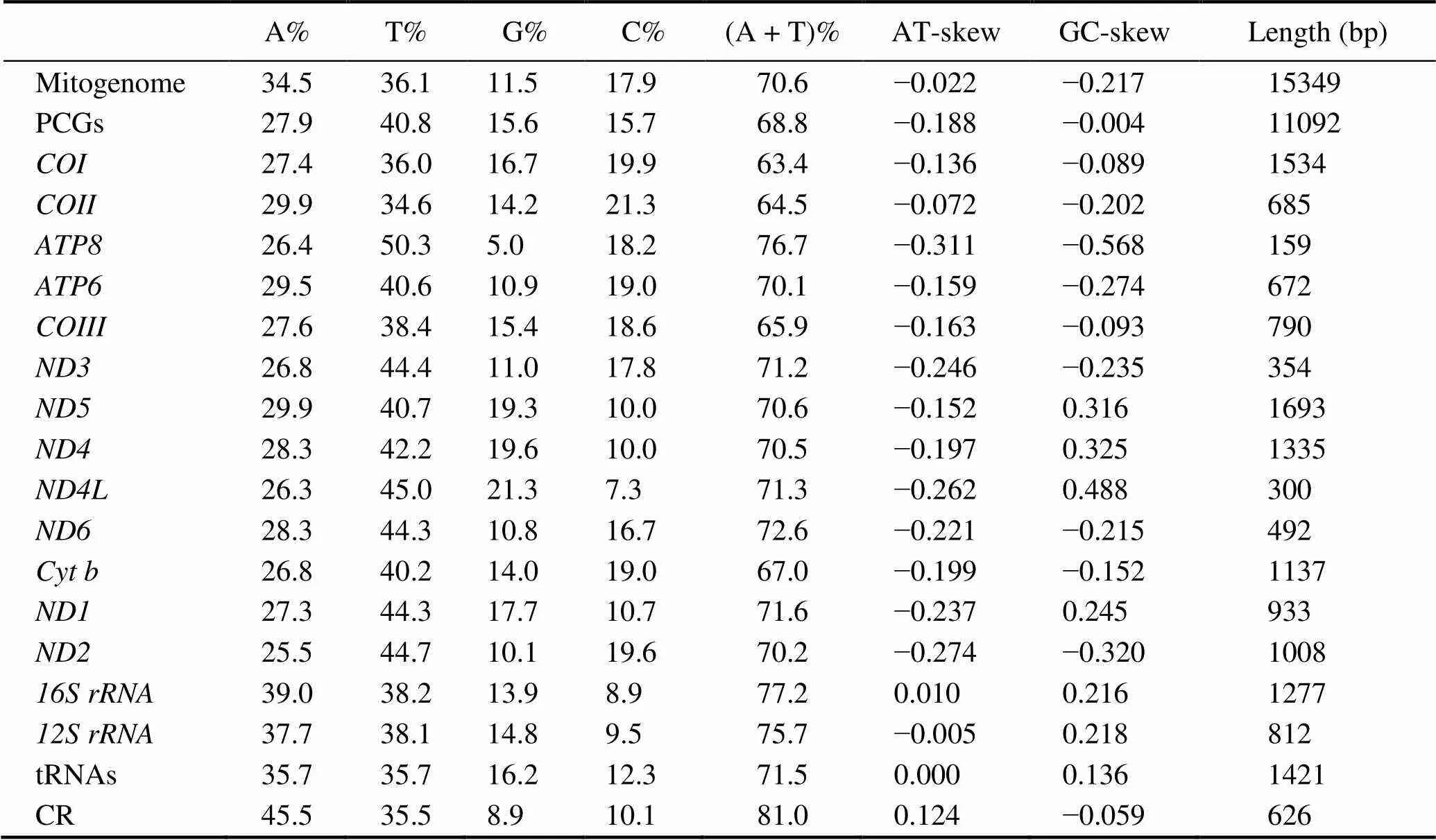
Table 3 Composition and skewness of the M. affinis mitogenome
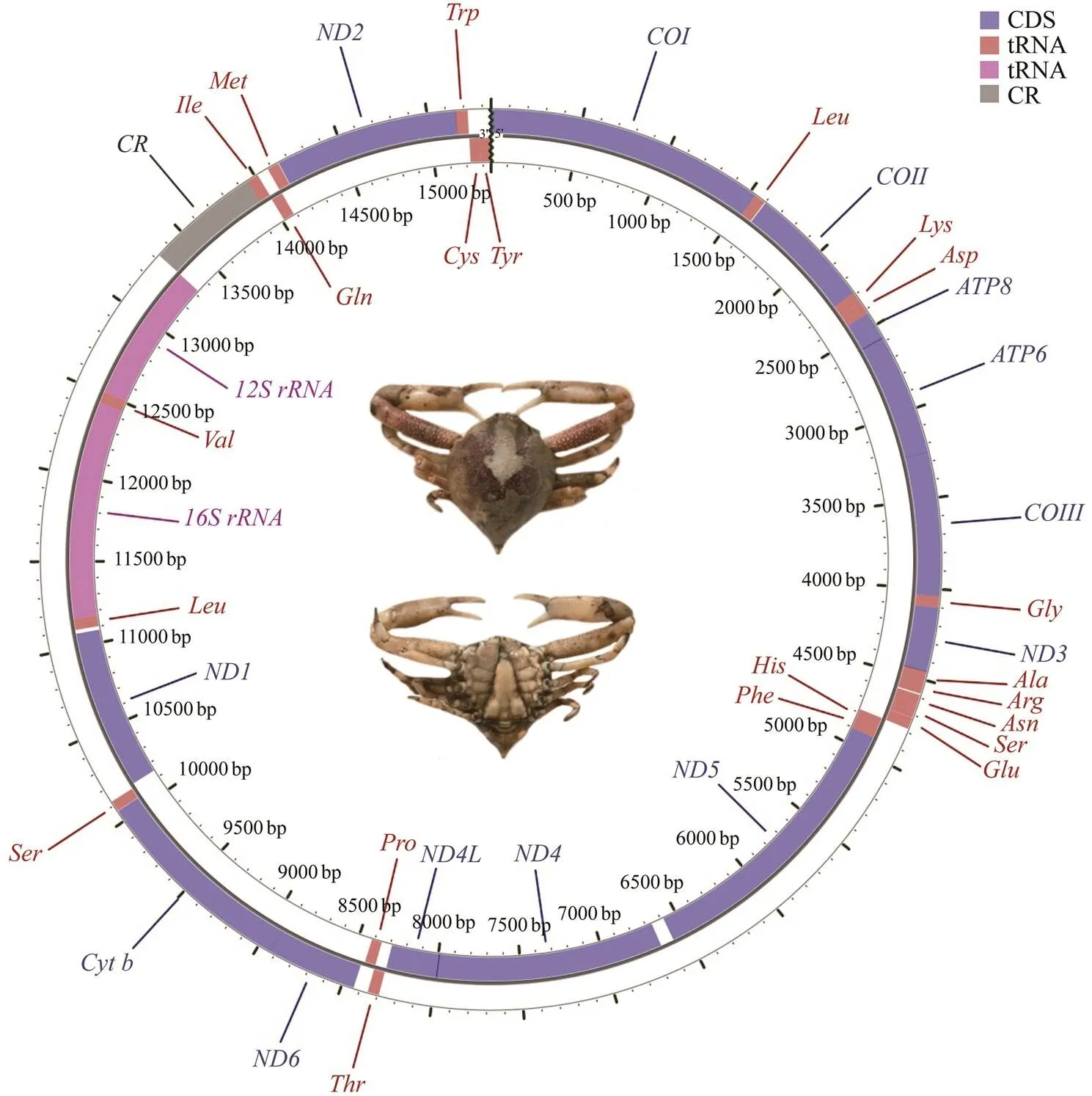
Fig.1 Gene map of the M. affinis mitogenome.
Among the 13 PCGs, the most frequently used amino acid are Leu (15.57%), Ser (10.31%), Phe (9.57%), and Ile(8.11%). In comparison, the least common amino acids are Cys (1.06%), Arg (1.63%), Gln (1.76%), and Asp (1.90%) (Fig.2A, Table 4). RSCU analysis shows that the most fre- quently used codons include UUA (Leu), UCU (Ser), and CCU (Pro); by contrast, GCG (Ala), AGC (Ser), CCG (Pro), and CGC (Arg) are the codons with the least frequencies (Fig.2B, Table 4). The preference for NNU and NNA co- dons can clearly be observed in the mitochondrial PCGs, which is consistent with the case of other Brachyuran spe- cies (Wang., 2020a; Wang., 2020b; Zhang., 2020b).
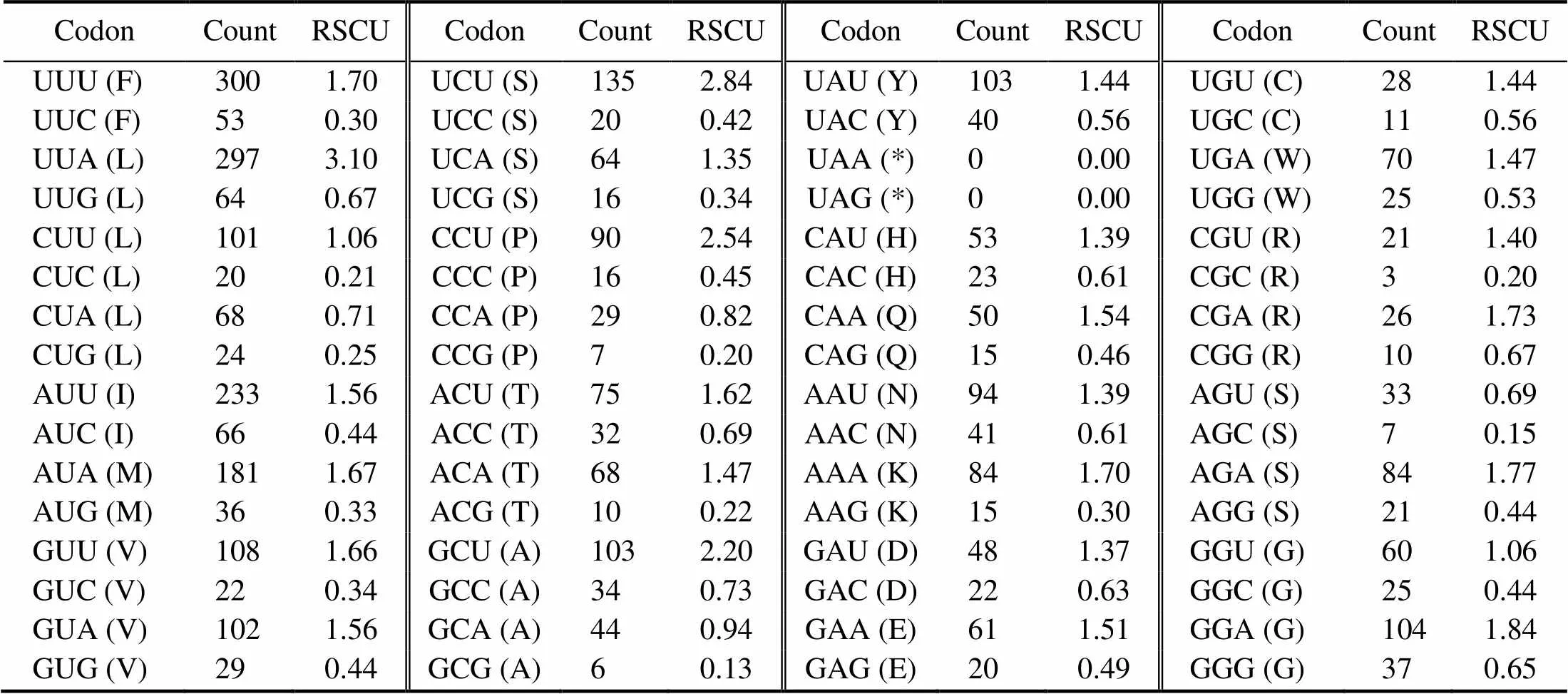
Table 4 Codon numbers and relative synonymous codon usage (RSCU) of 13 PCGs in the M. affinis mitogenome

Fig.2 Amino acid composition (A) and relative synonymous codon usage (B) of the M. affinis mitogenome.
3.3 Transfer RNAs (tRNAs), Ribosomal RNAs (rRNAs), and Control Region
A total of 22 tRNA genes, with lengths varying from 61bp to 70bp, are identified in the mitogenome of.Most tRNAs can fold into the typical cloverleaf-like struc- ture exceptfor-, which lacks the dihydrouridine arm (Fig.3). This feature seems common in metazoan mi- togenomes (Wang., 2015; Gong., 2019; Gong., 2020). In addition to the Watson-Crick base pairs (A–Tand G–C) and G–U matches, three types of mismatches are found in the stem region of tRNA. One C–U base pair is predicted in-, one A–A base pair is predicted in-, and three U–U base pairs are predicted in-,-,and-. Such stem mismatches ap- pear to be a common phenomenon in mitochondrial tRNA genes and may be corrected through post-transcriptional editing (Lavrov., 2000). The most reasonable expla- nation for this finding is that mitogenomes are unaffected by the recombination process. Therefore, base mismatches are able to exist, which may be helpful for eliminating de- leterious mutations (Lynch, 1997).
The(812bp) andgenes (1277bp) of themitogenome are typically separated by(Fig.1, Table 1). Both genes exhibit positive GC-skew (Table 3), thereby indicating that they are encoded by the L-strand. The CR of the mitogenome is located betweenand. The 626 bp-long CR is obvious- ly AT-biased (81.0%). The AT- and GC-skews are 0.124 and ?0.059; respectively (Table 3), thus indicating an obvious bias toward the use of As and Cs.
3.4 Phylogenetic Analysis
In the present study, the phylogenetic relationships among Brachyura were reconstructed according to the nucleotide sequences of 13 PCGs using the ML and BI methods.The ML tree and BI trees show an identical topology; thus, only one topology (., BI) with both support values is dis- playedin Fig.4. The results show thatandare most closely related among the species obtained and form a sister group at the end of the phylogenetic tree. Thus far, phylogenetic studies of Leucosiidae at the mole- cular level are extremely rare, and the evolutionary status of this family is unclear (Park., 2017). Although Park. (2017) presented the first and only research on the phylogenetic status of Leucosiidae using whole mitoge-nomes, the phylogenetic location of this family was not well resolved because of its limited representatives. By con-trast, the sister relationship between Leucosiidae and Matu-tidae in our phylogenetic trees is well supported (poste- rior probabilities=1; bootstrap=98). Considering that only two mitogenomes of this family have been analyzed, more sequences of leucosiid crabs are needed to be analyzed to reconfirm the phylogenetic position of Leucosiidae.
The monophyly of each of the 30 families included in this phylogeny, except Xanthidae and Homolidae, is strong- ly supported. Four Xanthidae species are divided into two clades, while three of these species cluster together as a clade, and the remaining species()forms a sister clade with the single representative of Ozii- dae ().Regarding the non-monophyly ofHomolidae, a single representative of Latreilliidae () forms a sister clade with a member ofHo- molidae (), thereby highlighting the autho- ritative identification ofand the necessity of col- lecting more representatives of Latreilliidae in future work.When viewed from a higher taxonomic level, our tree topo- logy indicates the existence of two major clades: section Dromiacea (the most basal clade) and the other two sections(Eubrachyura+Raninoida).The sister relationship betweenEubrachyura and Raninoida is strongly supported(poste- rior probabilities=1; bootstrap=100).The monophyly of Eubrachyura is well established (Von Sternberg and Cum berlidge, 2001; Tsang., 2014; Ma., 2019), while that of the other sections remains controversial due to li- mited mitogenomic data. Furthermore, because a third of the families (10/30) include only one representative, the non-monophyly of relevant families should be treated with some cautions. In the future studies, larger taxon samples are re- quired to resolve the origin and evolutionary relationships among Brachyura conclusively.
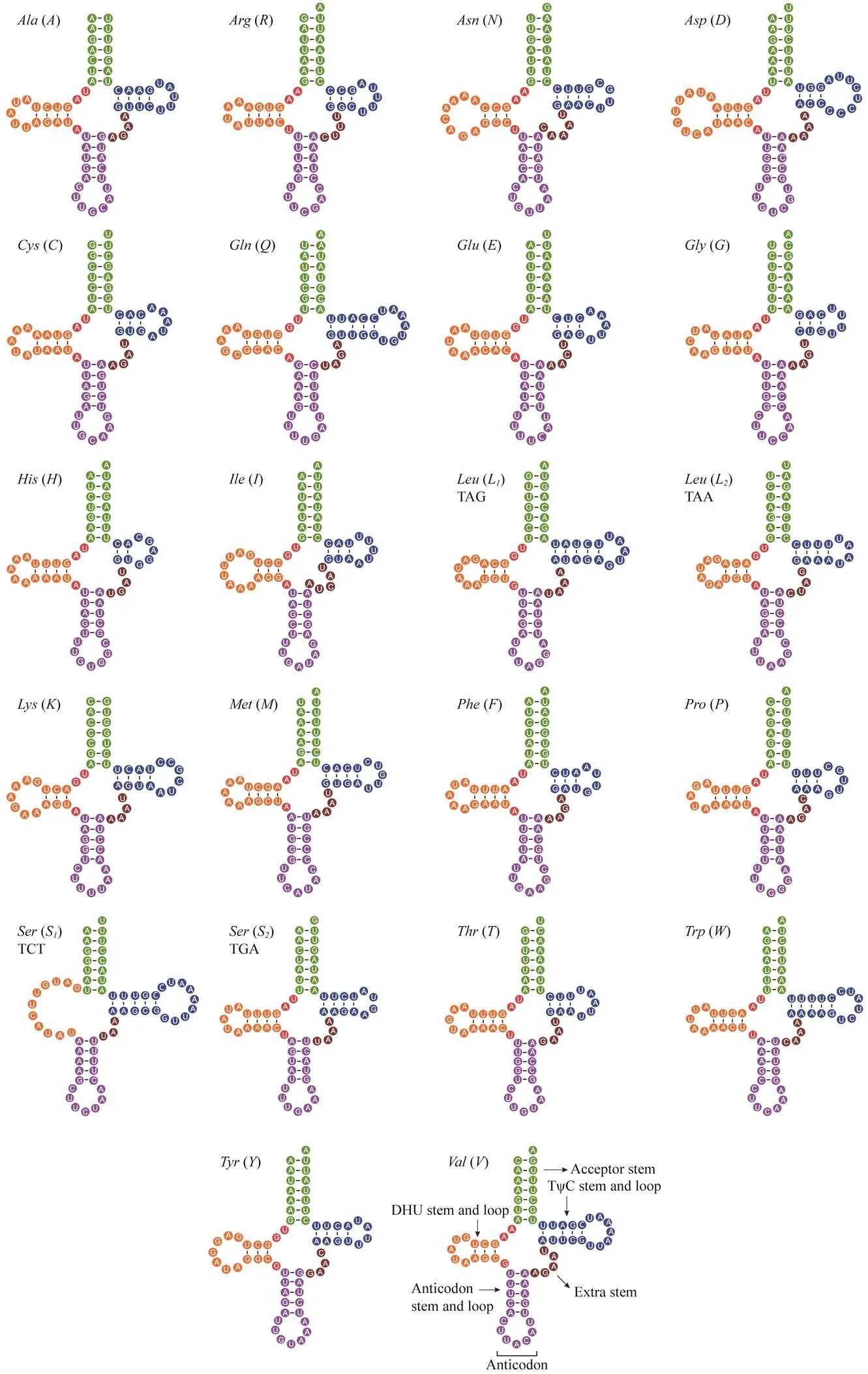
Fig.3 Potential secondary structures of 22 inferred tRNAs in the M. affinis mitogenome.
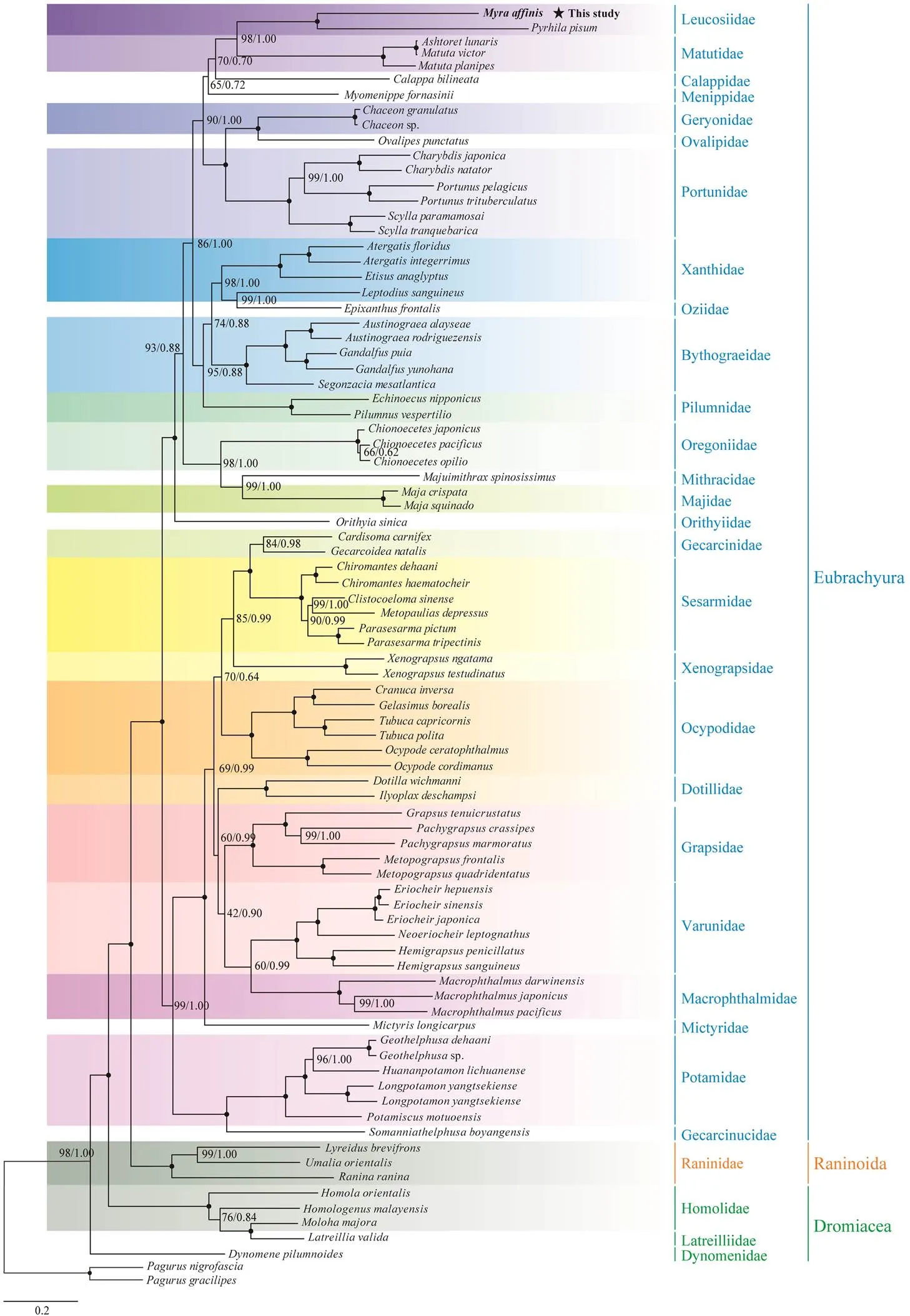
Fig.4 Phylogenetic tree of Brachyuran species inferred from the nucleotide sequences of 13 PCGs on the basis of maxi- mum likelihood (ML) and Bayesian inference (BI) analyses. Nodes marked with a solid circle indicate an ML bootstrap value of 100 and a supporting value of 100%.
4 Conclusions
In this study, we first determine and describe the com- plete mitogenome ofto supplement the limited mitogenome information of Leucosiidae.The 15349bp- long mitogenome contains 37 genes and an AT-rich region, which is typical in metazoan mitogenomes. All PCGs are initiated by the canonical start codon ATN and terminated by the TAN codon or an incomplete T.Phylogenetic ana-lyses support the sister relationship of Leucosiidae and Ma- tutidae on the basis of 13 PCGs. Furthermore, the mono- phyly of Eubrachyura is well established, and its sister re- lationship with Raninoida is strongly supported. Neverthe- less, comprehensive taxon sampling is necessary to resolve the phylogeny and origin of Brachyura with better accuracy.
Acknowledgements
This work was supported by the Natural Science Foun- dation of Zhejiang Province (No. LY21C190007) and the Zhoushan Science and Technology Bureau (No. 2021C21 007). We would like to express our gratitude to Dr. Jian Chen for helping in the species identification and provid- ing critical comments.
Ahyong, S. T., Lai, J. C., Sharkey, D., Colgan, D. J., and Ng, P. K., 2007. Phylogenetics of the brachyuran crabs (Crustacea: Decapoda): The status of Podotremata based on small subunit nuclear ribosomal RNA., 45 (2): 576-586.
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G.,., 2013. MITOS: Improvedmetazoanmitochondrial genome annotation., 69 (2): 313-319.
Boore, J. L., 1999. Animal mitochondrial genomes., 27 (8): 1767-1780.
Chen, J., Xing, Y., Yao, W., Zhang, C., Zhang, Z., Jiang, G.,., 2018. Characterization of four new mitogenomes from Ocy- podoidea & Grapsoidea, and phylomitogenomic insights into thoracotreme evolution., 675: 27-35.
Davie, P. J., Guinot, D., and Ng, P. K., 2015.In:–Koninklijke Brill NV, Leiden, 921-979.
De Melo, G. A. S., 1996..Plêiade/FAPESP, S?o Paulo, 603-604.
Dierckxsens, N., Mardulyn, P., and Smits, G., 2017. NOVOPlasty:assembly of organelle genomes from whole genome data., 45 (4): e18-e18.
Galil, B., 2001. A revision ofLeach, 1817 (Crustacea: De- capoda: Leucosioidea)., 75: 409- 446.
Gong, L., Liu, B., Liu, L., Guo, B., and Lü, Z., 2019a. The com- plete mitochondrial genome of(Centrarchifor- mes: Terapontidae) and comparative analysis of the control re- gion among eight Centrarchiformes species., 45 (2): 137-144
Gong, L., Lu, X., Wang, Z., Zhu, K., Liu, L., Jiang, L.,.,2019b. Novel gene rearrangement in the mitochondrial genome of(Anomura: Coenobitidae) and phylo-genetic implications for Anomura., 112 (2): 1804- 1812.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A.,and Jermiin, L. S., 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates., 14 (6): 587- 589.
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T., 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform., 30 (14): 3059- 3066.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K., 2018. MEGA X: Molecular evolutionary genetics analysis across com-puting platforms., 35 (6): 1547-1549.
Kumar, V., Tyagi, K., Chakraborty, R., Prasad, P., Kundu, S., Tyagi, I.,., 2020. The complete mitochondrial genome of endemic giant tarantula,(Araneae: The- raphosidae) and comparative analysis., 10 (1): 1-11.
Lavrov, D. V., Brown, W. M., and Boore, J. L., 2000. A novel typeof RNA editing occurs in the mitochondrial tRNAs of the cen- tipede Lithobius forficatus., 97 (25): 13738-13742.
Lowe, T. M., and Chan, P. P., 2016. tRNAscan-SE on-line: Integrating search and context for analysis of transfer RNA genes., 44 (W1): W54-W57.
Lu, X., Gong, L., Zhang, Y., Chen, J., Liu, L., Jiang, L.,., 2020. The complete mitochondrial genome of: The first representative from the family Calappidae and its phylogenetic position within Brachyura., 112 (3): 2516-2523.
Lynch, M., 1997. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes., 14 (9): 914-925.
Ma, K. Y., Qin, J., Lin, C. W., Chan, T. Y., Ng, P. K., Chu, K. H.,., 2019. Phylogenomic analyses of brachyuran crabs sup- port early divergence of primary freshwater crabs., 135: 62-66.
Martin, J. W., and Davis, G. E., 2001.. Natural History Museum of Los An- geles County, Los Angeles, 1-124.
Martin, M., 2011. Cutadapt removes adapter sequences from high- throughput sequencing reads., 17 (1): 10-12.
Naderloo, R., 2017. FamilyLeucosiidae Samouelle, 1819 (pebble crabs).In:. Springer, Cham, 75-120.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q., 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies., 32 (1): 268-274.
Ojala, D., Montoya, J., and Attardi, G., 1981. tRNA punctuation model of RNA processing in human mitochondria., 290(5806): 470-474.
Park, Y. J., Park, C. E., Jung, B. K., Ibal, J. C., Jung, Y., Hong, S. J.,., 2017. The first complete mitochondrial genome sequence of the leucosiid crab(Arthropoda, Decapoda, Leucosiidae)., 2 (2): 885- 886.
Perna, N. T., and Kocher, T. D., 1995. Patterns of nucleotide com- position at fourfold degenerate sites of animal mitochondrial genomes., 41 (3): 353-358.
Rocha, C. T., Regina, W. M., Mantelatto, F. L., Christopher, T., and José, Z. F., 2018. Ultrastructure of spermatozoa of members of Calappidae, Aethridae and Menippidae and discussion of theirphylogenetic placement., 101 (1): 89-100.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Dar- ling, A., Hohna, S.,., 2012. MrBayes 3.2: Efficient Baye- sian phylogenetic inference and model choice across a large model space., 61 (3): 539-542.
Ruan, H., Li, M., Li, Z., Huang, J., Chen, W., Sun, J.,.,2020. Comparative analysis of complete mitochondrial genomes of tThreefishes (Perciformes: Gerreidae) and primary exploration of their evolution history., 21 (5): 1874.
Sanchez, G., Tomano, S., Yamashiro, C., Fujita, R., Wakabayashi, T., Sakai, M.,., 2016. Population genetics of the jumbo squid(Cephalopoda: Ommastrephidae) in the northern Humboldt Current system based on mitochondrial and microsatellite DNA markers., 175: 1-9.
Schweitzer, C. E., and Feldmann, R. M., 2010. The oldest Brach-yura (Decapoda: Homolodromioidea: Glaessneropsoidea) known to date (Jurassic)., 30 (2): 251- 256.
Spears, T., Abele, L. G., and Kim, W., 1992. The monophyly of Brachyuran crabs: A phylogenetic study based on 18S rRN?., 41 (4): 446-461.
Stothard, P., and Wishart, D. S., 2005. Circular genome visuali- zation and exploration using CGView., 21 (4): 537-539.
Talavera, G., and Castresana, J., 2007. Improvement of phyloge- nies after removing divergent and ambiguously aligned blocks from protein sequence alignments., 56 (4): 564-577.
Tan, C., 1996. Leucosiidae of the Albatross expedition to the Phi- lippines, 1907–1910 (Crustacea: Brachyura: Decapoda)., 30 (7): 1021-1058.
Tan, M. H., Gan, H. M., Lee, Y. P., Linton, S., Grandjean, F., Bartholomei-Santos, M. L.,., 2018. ORDER within the chaos: Insights into phylogenetic relationships within the Ano- mura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements., 127: 320-331.
Tsang, L. M., Schubart, C. D., Ahyong, S. T., Lai, J. C., Au, E. Y.,Chan, T. Y.,., 2014. Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origin of freshwater crabs., 31 (5): 1173-1187.
Von Sternberg, R., and Cumberlidge, N., 2001. On the hetero- treme-thoracotreme distinction in the Eubrachyura de Saint Laurent, 1980 (Decapoda, Brachyura)., 74 (4): 321- 338.
Wang, Q., Tang, D., Guo, H., Wang, J., Xu, X., and Wang, Z., 2020a. Comparative mitochondrial genomic analysis ofand insights into the phylogeny of the Ocypo- doidea & Grapsoidea., 112 (1): 82-91.
Wang, X., Huang, Y., Liu, N., Yang, J., and Lei, F., 2015. Seven complete mitochondrial genome sequences of bushtits (Passe- riformes, Aegithalidae,): The evolution pattern in dup- licated control regions., 26 (3): 350-356.
Wang, Z., Shi, X., Guo, H., Tang, D., Bai, Y., and Wang, Z., 2020b.Characterization of the complete mitochondrial genome ofand comparison with other Brachyuran crabs., 112 (1): 10-19.
Zhang, D., Gao, F., Jakovlic, I., Zou, H., Zhang, J., Li, W. X.,.,2019. PhyloSuite: An integrated and scalable desktop platformfor streamlined molecular sequence data management and evo-lutionary phylogenetics studies.,20 (1): 348-355.
Zhang, Y., Gong, L., Lu, X., Jiang, L., Liu, B., Liu, L.,., 2020.Gene rearrangements in the mitochondrial genome of(Brachyura: Sesarmidae) and phylogenetic im- plications for Brachyura., 162: 704-714.
Zhang, Z. Q., 2011.. Magno- lia Press, Auckland, 1-237.
December 21, 2020;
February 16, 2021;
July 20, 2021
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
.E-mail:lzmnblzm@163.com
(Edited by Qiu Yantao)
 Journal of Ocean University of China2022年4期
Journal of Ocean University of China2022年4期
- Journal of Ocean University of China的其它文章
- Multisource Target Classification Based on Underwater Channel Cepstral Features
- Joint Model of Wind Speed and Corresponding Direction Based on Wind Rose for Wind Energy Exploitation
- Elastic-Wave Reverse Time Migration Random Boundary-Noise Suppression Based on CycleGAN
- Identification, Phylogeny and Expressional Profiles of Peptidoglycan Recognition Protein (PGRP) Gene Family in Sinonovacula constricta
- Molecular Characterization,Expression Pattern and Transcriptional Regulation of Figla During Gonad Development in Japanese Founder(Paralichthys olivaceus)
- Expressions of Toll Like Receptor (TLR) Genes in Paralichthys olivaceus After Induced by Different Extracts of Edwardsiella tarda
