ldentification and expression of the CEP gene family in apple(Malus×domestica)
Ll Rui, AN Jian-ping, YOU Chun-xiang, SHU Jing, WANG Xiao-fei, HAO Yu-jin
1 National Key Laboratory of Crop Biology, National Research Center for Apple Engineering and Technology/College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an 271018, P.R.China
2 Shandong Agriculture and Engineering University, Jinan 251100, P.R.China
1. lntroduction
The majority of signaling peptides are small cleavage products of precursor peptides. Several of these precursors must be post-translationally modified to form mature peptides, which usually contain 20 amino acids (Butenkoet al.2009; Murphyet al. 2012). In theArabidopsisgenome, over 1 000 putative small signaling peptides have been predicted,and some of them have been functionally identified, including C-TERMINALLY ENCODED PEPTIDE1 (CEP1), CLAVATA3(CLV3), CLV3/EMBRYO SURROUNDING REGIONRELATED (CLE), and RAPID ALKALINIZATION FACTOR (RALF)(Czyzewiczet al. 2013). Small and secreted regulatory peptides are a growing class of signaling molecules that are involved in regulating plant developmental programs and adapting to extreme environmentviacell-to-cell communica-tion (Katsiret al. 2011; Murphyet al. 2012). Several families of regulatory peptides have been functionally identified and have been found to participate in this process (Butenkoet al. 2009;Czyzewiczet al. 2013; Leeet al. 2015; Songet al. 2016).
Among the peptide families, the C-TERMINALLYENCODED PEPTIDE (CEP) genes contain a conserved 15-amino acid peptide domain at or near the C-terminus and are characterized inArabidopsis(Ohyamaet al. 2008). The post-translationally modified CEP family members contain an N-terminal secretion signal (NSS) and one or more conserved CEP domains (Mohd-Radzmanet al. 2015).CEPgenes are widely present among gymnosperm and angiosperm plants, but absent in land plants that lack true branching roots or root vasculature, indicating that their emergence coincides with the evolution of seed plants(Delayet al. 2013; Ogilvieet al. 2014). Members of theCEPfamily have already shown to regulate plant lateral root and root nodule development as well as root/shoot growth(Delayet al. 2013; Robertset al. 2013; Mohd-Radzman and Laffont 2016). For example, overexpression ofAtCEP1or treatment with chemically synthesized CEP1 peptide inArabidopsisresults in a reduction in the number of emerged lateral roots and the inhibition of primary root growth (Ohyamaet al. 2008). InMedicago, overexpression ofMtCEP1increases nodulation by promoting rhizobial infections and exhibits repression in lateral root development (Iminet al.2013; Mohd-Radzman and Laffont 2016). In addition,CEPgenes are reported to be negative regulators that mediate environmental influences on plant development (Delayet al. 2013). TheAtCEP3loss-of-function mutant enhances root development under adverse environmental conditions(Delayet al. 2013).
Currently, an enormous number of possible peptide ligand-receptors (kinases) has been identified and several receptor-like proteins such as CLV1/2, FER, and CEPR1/2 have been found to be involved with root development through interaction with peptides (Kondoet al. 2011; Duet al. 2016; Robertset al. 2016). The leucine-rich repeat(LRR) receptor kinases CEP RECEPTOR 1 (CEPR1;At5g49660) and CEP RECEPTOR 2 (CEPR2; At1g72180)have been shown to be the receptor for CEP1 and other CEPs (Bryanet al. 2012; Tabataet al. 2014; Robertset al.2016). Further studies may reveal that CEPs and CEPR1 participate in the N-dependent responses in long-distance systemic signaling pathways (Okamotoet al. 2016).
Apple (Malus×domestica) is one of the most widely cultivated fruit crops worldwide and is the most economically important woody plant in temperate regions (Dimick and Hoskin 1983; Leeet al. 2007). The draft genome sequence of apple has been completed, which allows genome-wide analyses of specific gene families (Velascoet al. 2010). Genome-wide analyses of theCEPgenes have been reported inArabidopsis thaliana(Delayet al. 2013) andOryza sativa(Suiet al. 2016), and several gene families have been identified in apple. However, there is no genome wide data regarding the appleCEPgenes. In brief, small signaling peptides play an essential role in all stages of plant growth and development.The characterization of apple CEP peptides provides insight into the molecular mechanism of apple root growth and the responses to different environmental factors.
In this study, a genome-wide analysis of theCEPgene family was conducted using the apple genome database,and the chromosome locations and gene structures of the putativeCEPgenes were analyzed. Next, the motif compositions of theMdCEPswere obtained using the MEME Program. Subsequently, the expression patterns ofMdCEPgenes in different tissues and in response to abiotic stresses were analyzed. This study provides a foundation for future research into the functional roles ofMdCEPgenes.
2. Materials and methods
2.1. ldentification and annotation MdCEP genes in apple
To identify members of theCEPgene family, all knownArabidopsisCEP protein sequences were retrieved from the database of Institute for Genomic Research (TIGR) and used as queries in BLASTP searches against the Genome Database for Rosaceae (GDR) (http://www.rosaceae.org/).Stand-alone versions of BLASP (http://blast.ncbi.nlm.nih.gov), which are available from NCBI, were used with the e-value cutoff of 1e-003. Then, the predictedCEPgene family sequences were downloaded from the GDR database. All of the protein sequences that were derived from the selectedMdCEPcandidate genes were examined with the domain analysis programs Pfam (http://pfam.sanger.ac.uk/) and Simple Modular Architecture Research Tool(SMART; http://smart.embl-heidelberg.de/), with the default cutoff parameters.
2.2. Chromosomal locations and gene structures of MdCEP genes
The chromosomal locations and gene structures were retrieved from the apple genome data that were downloaded from the GDR database. The chromosomal map showing the physical location of all of theMdCEPgenes was generated with the MapDraw Software and the gene structures of theMdCEPgenes were generated with GSDS (http://gsds.cbi.pku.edu.cn/). The isoelectric point (pI) and molecular weight of MdCEPs were obtained with the assistance of proteomics and sequence analysis tools on the ExPASy Proteomics Server (http://expasy.org/). All putative MdCEPs were analyzed by Multiple Em for Motif Elicitation (MEME, ver.4.11.2), a motif search tool to identify conserved motifs shared among CEP proteins with the following parameters, optimum motif width ranges from 6 to 20 and a maximum number of motifs of 10. All other parameters were set at default.
2.3. Sequence alignment and phylogenetic analysis
Multiple sequence alignments of the MdCEP sequences(MdCEP1-27) and AtCEP sequences (AtCEP1-15) were performed using Clustal X (ver. 2.1). The MUSCLE Program(ver. 3.52) was also used to perform multiple sequence alignments to confirm the Clustal X result (Edgar 2004).Phylogenetic trees of the MdCEPs were constructed based on the protein sequences according to the neighbor-joining(NJ) method of the MEGA5 Program (ver. 5.1, http://www.megasoftware.net/) (Wanget al. 2012; Hall 2013), with p-distance and the complete deletion option parameters engaged. The reliability of the trees was tested using bootstrapping with 1 000 replicates. The images of the phylogenetic treeos were drawn using the MEGA5 Program.
2.4. Plant materials, treatments and gene expression analysis
To investigate the expression ofMdCEPgenes under different apple tissues, the samples (roots, stems, leaves, flowers and fruits) from a five-year-old apple tree were collected.In vitroshoot cultures of apple Gala were maintained on Murashige and Skoog (MS) medium containing 0.5 mg L–16-BA, 0.2 mg L–1NAA, and 0.1 mg L–1gibberellin (GA)under long-day conditions (16 h light/8 h dark cycle) and then subcultured with a 4 weeks interval (Anet al. 2016).Two-week-old apple shoot cultures were treated separately with either 0.5 mmol L–1nitrates, 200 mmol L–1NaCl, 300 mmol L–1mannitol, 10% PEG, 4°C temperature, or 100 μmol L–1ABA. Untreated apple tissue cultures were used as controls. The samples were treated for 6 h before the expression analysis. All the samples were stored at ?80°C.RNA was extracted from triplicate biological replicates of the above samples using the Trizol method (Caoet al. 2013).All of the primers are shown in Appendix A. The results were based on the average of three parallel experiments.
3. Results
3.1. ldentification of CEP genes in apple
In total, 27 typicalCEPgenes were identified using theArabidopsisCEP domain as the unique identifier after manually removing the repeated and incomplete sequences from our data. These genes contained full length open reading frames (ORFs). Each gene was named based on its location on the chromosome (Table 1). The amino acids from the 89th(MdCEP26) to the 1 125th (MdCEP1) were encoded by the identifiedMdCEPgenes, which predicted that the molecular masses were 9.1-125.4 kDa and the pI of these proteins was between 5.58 (MdCEP3) and 10.62 (MdCEP25).
3.2. Phylogenetic tree of the apple CEP proteins
To evaluate the evolutionary relationships among the MdCEP proteins, the phylogenetic analysis was performed for theArabidopsis(15 members) and apple (27 members)CEP proteins using NJ method of the MEGA5 Program(Fig. 1). Next, the unrooted phylogenetic tree was constructed using the multiple sequence alignment file. The phylogenetic tree of the MdCEPs was divided into three classes (I, II and III) of monophyletic clades and 7 sister pairs of paralogousMdCEPgenes were found with strong bootstrap support (>90%). In addition, Class I contained sixMdCEPgenes. Class III contained 10MdCEPgenes, and class II constituted the largest clade, with 11MdCEPgenes.The CEP protein in the same group has a similar amino acid sequence of the CEP domain (Appendix B).
3.3. Chromosomal localizations of CEPs in the apple genomes
The chromosomal location for eachMdCEPgene was analyzed to visualize the relationships among genes. The results showed that the genes were distributed unevenly. Among these 27MdCEPgenes, threeMdCEPgenes (MdCEP25,MdCEP26andMdCEP27) were localized to unassembled genomic sequence scaffolds and could not be conclusively mapped to any chromosome. The remaining 24MdCEPgenes were present on 10 of 17 chromosomes (Fig. 2). Notably, chromosome 1 encompassed the largest quantity with sixMdCEPgenes, whereas chromosomes 10, 11 and 12 contained only oneMdCEPgene. For the remaining genes,threeMdCEPgenes were distributed on chromosomes 1,5 and 16, and two genes were located on chromosomes 2,6 and 8. Interestingly, threeMdCEPgenes (MdCEP1,Md-CEP2, andMdCEP3) and three pairs ofMdCEPs(MdCEP11andMdCEP12, MdCEP17andMdCEP18,MdCEP22andMdCEP23) were tightly colocalized on the apple genome. In this study, the large-scale segmental duplication events were investigated, which were thought to have occurred during the evolutionary process. Among theMdCEPgenes, a total of 7 genes (MdCEP6,MdCEP13,MdCEP14,MdCEP15,MdCEP16,MdCEP20, andMdCEP21) were found in the segmental duplication blocks. We also noted thatMdCEP6andMdCEP13were on the same pair of homologous chromosomes (Chr. 5 and Chr. 10) and thatMdCEP11,MdCEP12,MdCEP17,MdCEP18, andMdCEP19were on another pair of homologous chromosomes (Chr. 8 and Chr. 15). Additionally,they have been classified into the class II phylogenetic group.This confirms that genome duplications have contributed to gene family expansions throughout plant evolution (Cannonet al. 2004; Liet al. 2015).

Table 1 Information of C-TERMINALLYENCODED PEPTIDE (CEP) gene family in apple
3.4. Gene structure of CEP genes
To analyze the structural diversity and understand the possible structural evolution ofMdCEPgenes, the exon/intron organization in the coding sequences of the individualMdCEPgenes were analyzed. As illustrated in Fig. 3, the number of introns varied from 0 to 21 in theMdCEPgene family. Among the 27MdCEPgenes, there were 16 genes with no introns,and four genes (MdCEP11,MdCEP14,MdCEP20 andMd-CEP27) with only one intron. Unlike otherMdCEPs,MdCEP1,MdCEP2, andMdCEP3had many more introns. It should be noted that closely related members in the same subfamilies share similar gene length and exon/intron structure.
3.5. Protein motif analysis of the apple CEP gene family
To reveal the diversification of theMdCEPgenes, all of the MdCEP protein sequences were predicted by the MEME Program and 10 types of motifs were identified (Fig. 4). As expected, a common motif composition of the closely related members in the phylogenetic tree was revealed, suggesting functional similarities among the MdCEP proteins within the same subgroup (Fig. 4-A). The results showed that the CEP domain (motif 1) contained 15 amino acids and was shared by all members. In addition, three MdCEPs(MdCEP11, MdCEP12 and MdCEP19) were composed of more than five CEP domains; MdCEP14 and MdCEP17 had two CEP domains and the remaining MdCEPs had just one CEP domain (Fig. 4-B). Moreover, motif 5 existed in mostCEPgenes and it was predicted by the SignalP4.1 (http://www.cbs.dtu.dk/services/SignalP) to be the signal peptide which possibly contributed to the functions of CEP (Delayet al. 2013; Suiet al. 2016). Next, the WebLogo plots were assembled, and revealed that motifs 1 and 5 were highly conserved (Fig. 4-C), indicating that these sequences might have important biological functions.
3.6. Expression profiles of MdCEP genes in different tissues and in response to different abiotic conditions

Fig. 1 Phylogenetic relationship between Arabidopsis and apple C-TERMINALLYENCODED PEPTIDE (CEP) proteins. The protein sequences of the CEPs domain, including 27 from apple (MdCEPs, ★) and 15 from Arabidopsis (AtCEPs, ☆), were aligned by ClustalX (ver. 2.1) and were classified into 3 distinct classes (I, II and III). The phylogenetic tree was constructed using the neighborjoining (NJ) method by the MEGA (ver. 5.1) Software. The bootstrap values of 1 000 replicates were calculated at each node.
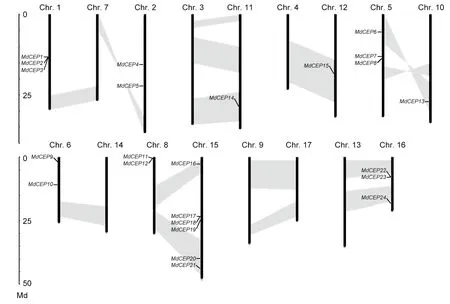
Fig. 2 Chromosome positions of C-TERMINALLYENCODED PEPTIDE (CEP) genes in apple (Malus×domestica, MdCEP). The chromosome number is indicated at the 10 of each chromosome representation. To simplify the presentation, we named the putative CEP family genes from MdCEP1 to MdCEP27 according to the gene order on chromosomes (MdCEP25, MdCEP26 and MdCEP27 were not found). Segmented duplicate homologous blocks are indicated with shadows.
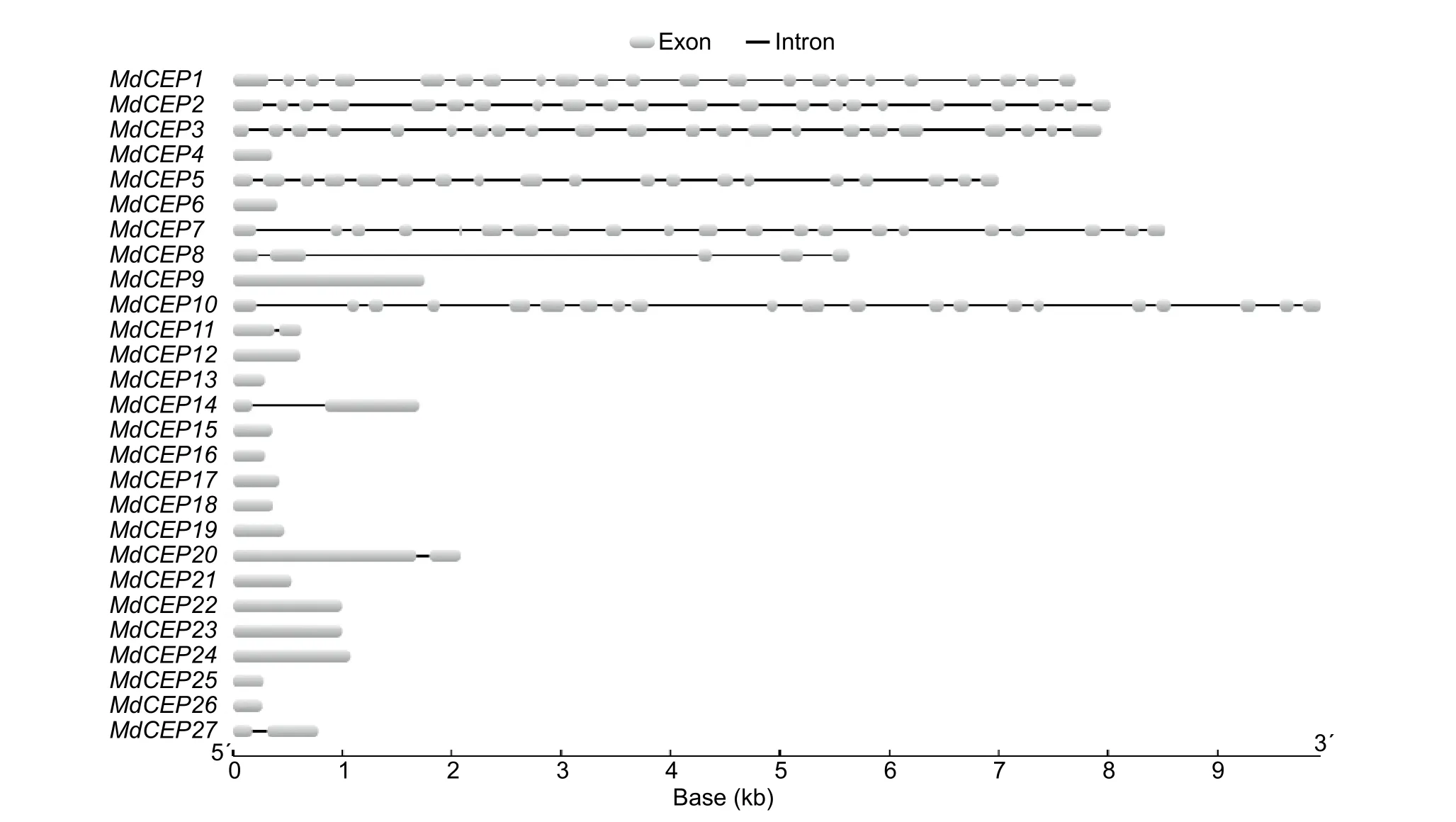
Fig. 3 Exon/intron structure of C-TERMINALLYENCODED PEPTIDE (CEP) genes in apple (Malus×domestica, MdCEP).
Gene expression patterns can provide important clues for gene functions (Kidokoroet al. 2015). The expression patterns ofMdCEPgenes were detected by quantitative real-time polymerase chain reaction (qRT-PCR) in five different organs (roots, stems, leaves, flowers, and fruits).The results showed that the tissue expression patterns of theCEPgenes were specific and expressed divergently(Fig. 5-A). Compared to other genes,MdCEP1,MdCEP6,MdCEP8andMdCEP16had a relatively high expression levels in stems, whereas,MdCEP9andMdCEP10were highly expressed in flowers and had lower levels of expression in fruits. It was noteworthy thatMdCEP14was mainly expressed in the leaves and thatMdCEP25was chiefly expressed in the fruits. Conversely, the transcript levels of the remainingMdCEPgenes were significantly higher in roots, withMdCEP22having the highest levels. Mounting evidence concludes thatCEPsexpression is regulated by environmental cues inArabidopsis(Delayet al. 2013). To examine whether the expression of appleMdCEPgenes were also induced by environmental cues and ABA treatment, the transcript abundance of all of theMdCEPgenes were tested under nitrogen limitation, salinity (NaCl) osmotic(Mannitol, PEG), cold (4°C) and exogenous ABA stress for 6 h. The results showed that expression ofMdCEPgenes were regulated by environmental factors and following ABA treatment (Fig. 5-B). The transcript levels ofMdCEP5,MdCEP6,MdCEP8andMdCEP10increased in response to the nitrogen limitation, while the expression of one-third ofMdCEPsgenes was decreased. The levels ofMdCEP1,MdCEP9,MdCEP13,MdCEP16andMdCEP27were enhanced in response to NaCl treatment. The expression ofMdCEP3,MdCEP5,MdCEP6,MdCEP8andMdCEP25was primarily induced by mannitol and PEG. MoreoverMdCEP15,MdCEP19andMdCEP20were significantly up-regulated under cold treatment. Notably,MdCEP17had the highest expression after ABA treatment. In addition, the genes within the same classes showed similar expression patterns. For example, members of the class II were induced by the ABA treatment. Most of theMdCEPgenes that were significantly up-regulated under low nitrogen conditions were classified as class III, while class I genes, includingMdCEP14,MdCEP21,MdCEP22, andMdCEP23,did not show significant changes. Interestingly, the expressions ofMdCEP21,MdCEP22,MdCEP23andMdCEP24were up-regulated by PEG treatment, but inhibited by the other treatments.

Fig. 4 Phylogenetic tree (A), protein motif analysis (B) and conserved domains (C) of C-TERMINALLYENCODED PEPTIDE (CEP)genes in apple (Malus×domestica, MdCEP). The amino acid sequences of the MdCEP proteins were aligned using Clustal X,and the phylogenetic tree was constructed using the neighbor-joining (NJ) method in the MEGA5 (ver. 5.1) Software. Schematic representation of the conserved motifs in the CEP proteins, which were elucidated using the MEME (ver. 4.11.4) Software. The black lines represent the unconserved sequences. The scale bar represents 200 aa.
4. Discussion

Fig. 5 Expression patterns of C-TERMINALLYENCODED PEPTIDE (CEP) genes of apple (Malus×domestica, MdCEP) in different organs (A) and under various abiotic conditions (B). Quantitative real-time polymerase chain reaction (qRT-PCR) was used to analyze the expression patterns of all the MdCEP genes in seedlings under the following conditions, nitrates (0.5 mmol L-1), salinity(NaCl, 200 mmol L–1), mannitol (300 mmol L–1), drought (PEG 6 000, 10% ), cold (4°C), and exogenous ABA (100 μmol L–1) for 6 h. The data were normalized to the apple actin expression level.
The CEPs, a class of small post-translationally modified signaling peptides, have been shown to be involved in regulating root architecture inArabidopsisand cell size in rice seeds (Robertset al. 2016; Suiet al. 2016). The genome-wide analyses of theCEPgene family have been reported inArabidopsisand rice. TheCEPfamily members displayed distinct expression patterns inArabidopsisdevelopment and were expressed divergently and exhibited tissue-specific expression patterns inOryza sativa(Delayet al. 2013; Suiet al. 2016). However, the biological functions for most CEPs remain largely unknown (Lee and De Smet 2016; Okamotoet al. 2016). In this study, twenty-sevenCEPgenes that contained the conserved CEP peptide domain were identified in apple. Then, a phylogenetic tree ofMdCEPsandAtCEPswas constructed and divided into three classes (class I, II and III) of monophyletic clades.Among them, the class III genes contained tenMdCEPs(but noAtCEPs), indicating that the appleCEPgenes have expanded during the evolutionary process (Cannonet al.2004). It is speculated thatCEPgenes may play a more complicated and important role in perennial woody plants(Delayet al. 2013; Robertset al. 2013). In addition, the gene structure ofCEPand the amino acid sequence of CEP domain in different groups were significantly different. For example, the CEP domain in class I contains more proline and glycine. The C-terminal GVG sequence of the CEP domain in class II is much conserved. The N-terminal part of most CEP domain (AYGQTGTGK) in class III showed high amino acid similarity.
The 27MdCEPgenes were localized to 10 of the 17 apple chromosomes. However, 3MdCEPgenes (MdCEP25,MdCEP26,andMdCEP27) were not conclusively mapped to any chromosome. It is hypothesized that otherMdCEPgenes exist in unknown genomic gaps, because only 81.3%of genomic sequences overlapped in the apple genome (Velascoet al. 2010; Wanget al. 2013). The protein motifs of the appleCEPgene family were analyzed using the MEME Program. As predicted, all MdCEPs contained more than one CEP domain (motif 1) and most MdCEPs contained motif 5 which may be the signal peptide. As shown in Fig. 4-C, the sequences of motifs 1 and 5 were highly conserved. Interestingly, the CEP domains inArabidopsisand apple both contain 15 amino acids and are highly conserved,which means that the genetic evolution of theCEPgenes structures is highly conserved in different plants.
Gene expression patterns provide important clues of gene function (Kidokoroet al. 2015). AllMdCEPgenes displayed different expression patterns, which represented the distinct roles of individualMdCEPgenes. In particularly,over two-thirds of theMdCEPgenes were highly expressed in the roots. It is hypothesized thatMdCEPgenes might have an indispensable role in regulating the growth and development of apple roots. Previous research found thatCEPgenes inhibit primary root length and lateral root number inArabidopsis(Robertset al. 2013; Robertset al. 2016).InMedicago, ectopic expression ofMtCEP1also reduces the lateral root number, but does not show any significant phenotypic variation after the knockdown ofMtCEP1alone.These data indicate thatCEPsare subject to functional redundancy (Iminet al. 2013; Mohd-Radzmanet al. 2015).
Plants are constantly under multiple abiotic stresses during their lifespan and have evolved unique strategies to integrate these cues into growth regulation to adapt to adverse environments (Glazebrook 2001; Chenet al. 2002;Zhaoet al. 2012; Suet al. 2013). Research has showed that the small secreted regulatory CEPs play prominent roles in adapting to adverse environmental conditions (Delayet al.2013). Recent studies indicate that CEPs are negative regulators of plant development and that over-expression ofCEPgenes occurs under nutrient or abiotic stress, particularly under nitrogen limitation (Tabataet al. 2014). In this study, we explored the expression ofMdCEPgenes under various growth conditions including nitrogen limitation, NaCl,mannitol, PEG, cold (4°C) and exogenous ABA. The results showed thatMdCEPgenes were induced to varying degrees under different treatments. Notably, nearly two-thirds ofMdCEPgenes were involved in the low nitrogen response,which is consistent with previous reports inArabidopsisandMedicago(Delayet al. 2013; Iminet al. 2013; Tabataet al.2014). Presumably these genes may have similar functions on vegetative growth in plants. MostMdCEPgenes were also significantly up-regulated under NaCl, mannitol, PEG and ABA treatments, indicating thatMdCEPgenes might participate in the drought stress response. Interestingly,the transcript levels ofMdCEP15,MdCEP19andMdCEP20were significantly higher under cold treatment, indicating that these genes may have similar functions in regulating the response to cold stress. These functions are achieved by interacting with membrane-bound receptors on target cells(Etchells and Turner 2010; Hirakawaet al. 2010; Okamotoet al. 2016; Robertset al. 2016). In addition, someMdCEPgenes are associated withAtCEPgenes in the same evolutionary branch and have similar expression patterns. For example, bothMdCEP26andAtCEP5respond significantly to nitrogen limitation.MdCEP25andAtCEP4have a significant response to mannitol treatment (Delayet al. 2013).
In this study, a bioinformatics approach was applied to perform a genome-wide survey ofCEPgenes in apple. Our data suggest that the 27 CEP proteins of apple form three subgroups according to their CEP domain. In addition, theCEPgene in the same subfamily has similar gene structure and amino acid sequence of CEP domain. The expression patterns ofMdCEPgenes offer a comprehensive framework to analyze the important role of the CEP peptides in plant growth and development. Our results show that the CEP domain sequences may influence gene function. For exam-ple, the expression ofCEPgene in roots was significantly higher in class I. Meanwhile, theMdCEPgenes of class I were induced by PEG treatment specifically. These results constitute the basis for further exploration of the mechanism of theCEPgene family, and indicate that the CEP domain is directly related to the biological function of theCEPgene with higher plants. In addition, peptide signals can be matched to their corresponding receptor through a variety of methods (Murphyet al. 2012). However, only very few peptide ligand-receptors have been shown to pair with corresponding peptides among a large number of possible peptide ligand-receptors (kinases) (Butenkoet al.2009; Songet al. 2016). Additionally, it was shown that one receptor-like protein kinase (RLK) can bind several signaling molecules and that some receptors may functionally replace one another (Murphyet al. 2012). It is unknown why and how these proteins act interchangeably. What we do know regarding peptide signaling potential, including the interaction with receptors and any downstream changes is just the tip of the iceberg. There are still many mysteries that need to be solved.
5. Conclusion
In this study, 27 appleCEPgenes were classified. Additionally, we presented a complete analysis of theMdCEPgene family utilizing the apple genome database. Next,qRT-PCR was used to determine the spatial expression ofMdCEPgenes, which exhibited tissue-specific expression patterns. Additionally, the expression level ofMdCEPgenes was increased by nitrogen limitation and a variety of other abiotic stresses. These data may facilitate the functional characterization of theCEPgene family.
Acknowledgements
We thank the earmarked fund for the China Agriculture Research System (CARS-28), the National Natural Science Foundation of China (31471854, 31601742), the earmarked fund for the Shandong Province Agriculture Research,China (SDAIT-06-03), the Natural Science Foundation of Shandong Province, China (ZR2011CQ007).
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
An J P, Li H H, Song L Q, Su L, Liu X, You C X, Hao Y J. 2016.The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple.Plant Physiology and Biochemistry, 108, 24-31.
Bryan A C, Obaidi A, Wierzba M, Tax F E. 2012. XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development inArabidopsis thaliana.Planta, 235, 111-122.
Butenko M A, Vie A K, Brembu T, Aalen R B, Bones A M. 2009.Plant peptides in signalling, looking for new partners.Trends in Plant Science, 14, 255-263.
Cannon S B, Mitra A, Baumgarten A, Young N D, May G. 2004.The roles of segmental and tandem gene duplication in the evolution of large gene families inArabidopsis thaliana.BMC Plant Biology, 4, 10.
Cao Z H, Zhang S Z, Wang R K, Zhang R F, Hao Y J. 2013.Genome wide analysis of the apple myb transcription factor family allows the identification ofMdoMYB121gene confering abiotic stress tolerance in plants.PLOS ONE,8, e69955.
Chen W, Provart N J, Glazebrook J, Katagiri F, Chang H S,Eulgem T, Mauch F, Luan S, Zou G, Whitham S A, Budworth P R, Tao Y, Xie Z, Chen X, Lam S, Kreps J A, Harper J F,Ammour A S, Mani B M, Heinlein M, Kobayashi K, Hohn T,Dangl J L, Wang X, Zhua T,et al. 2002. Expression profile matrix ofArabidopsistranscription factor genes suggests their putative functions in response to environmental stresses.The Plant Cell, 14, 559–574.
Czyzewicz N, Yue K, Beeckman T, De Smet I. 2013. Message in a bottle, small signalling peptide outputs during growth and development.Journal of Experimental Botany, 64,5281–5296.
Delay C, Imin N, Djordjevic M A. 2013.CEPgenes regulate root and shoot development in response to environmental cues and are specific to seed plants.Journal of Experimental Botany, 64, 5383–5394.
Dimick P S, Hoskin J C. 1983. Review of apple flavor - state of the art.Critical Reviews in Food Science and Nutrition,18, 387–409.
Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J. 2016. Receptor kinase complex transmits RALF peptide signal to inhibit root growth inArabidopsis.Proceedings of the National Academy of Sciences of the United States of America, 113, 8326–8334.
Edgar R C. 2004. MUSCLE, multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Research,32, 1792–1797.
Etchells J P, Turner S R. 2010. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division.Development, 137,767–774.
Glazebrook J. 2001. Genes controlling expression of defense responses inArabidopsis- 2001 status.Current Opinion in Plant Biology, 4, 301–308.
Hall B G. 2013. Building phylogenetic trees from molecular data with MEGA.Molecular Biology and Evolution, 30,1229–1235.
Hirakawa Y, Kondo Y, Fukuda H. 2010. Establishment and maintenance of vascular cell communities through local signaling.Current Opinion in Plant Biology, 14, 17–23.
Imin N, Mohd-Radzman N A, Ogilvie H A, Djordjevic M A. 2013.The peptide-encodingCEP1gene modulates lateral root and nodule numbers inMedicago truncatula.Journal of Experimental Botany, 64, 5395–5409.
Katsir L, Davies K A, Bergmann D C, Laux T. 2011. Peptide signaling in plant development.Current Biology, 21,356–364.
Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K,Mizoi J, Myint P S H N, Fujita Y, Sekita S, Shinozaki K. 2015.Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression.The Plant Journal, 81, 505–518.
Kondo Y, Hirakawa Y, Kieber J J, Fukuda H. 2011. CLE peptides can negatively regulate protoxylem vessel formationviacytokinin signaling.Plant & Cell Physiology, 52, 37–48.
Lee J S, De Smet I. 2016. Fine-tuning development through antagonistic peptides, an emerging theme.Trends in Plant Science, 21, 991–993.
Lee J S, Hnilova M, Maes M, Lin Y C, Putarjunan A, Han S K,Avila J, Torii K U. 2015. Competitive binding of antagonistic peptides fine-tunes stomatal patterning.Nature, 522,439–443.
Lee S Y, Chang S S, Shin J H, Kang D H. 2007. Membranefiltration method for enumeration and isolation ofAlicyclobacillusspp. from apple juice.Letters in Applied Microbiology, 45, 540–546.
Li X, Yin X, Wang H, Li J, Guo C. 2015. Genome-wide identification and analysis of the apple (Malus×domesticaBorkh.)TIFYgene family.Tree Genetics & Genomes, 11,1–13.
Mohd-Radzman N A, Binos S, Truong T T, Imin N, Mariani M,Djordjevic M A. 2015. Novel MtCEP1 peptides producedin vivodifferentially regulate root development inMedicago truncatula.Journal of Experimental Botany, 66, 5289–5300.
Mohd-Radzman N A, Laffont C. 2016. Different Pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development.Plant Physiology,171, 2536–2548.
Murphy E, Smith S, De Smet I. 2012. Small signaling peptides inArabidopsisdevelopment, how cells communicate over a short distance.The Plant Cell, 24, 3198–3217.
Ogilvie H A, Imin N, Djordjevic M A. 2014. Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes.BMC Genomics, 15, 870.
Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide inArabidopsisbyin silicogene screening, followed by LC-MS-based structure analysis.The Plant Journal, 55, 152–160.
Okamoto S, Tabata R, Matsubayashi Y. 2016. Long-distance peptide signaling essential for nutrient homeostasis in plants.Current Opinion in Plant Biology, 34, 35–40.
Roberts I, Smith S, De Rybel B, Van Den Broeke J, Smet W, De Cokere S, Mispelaere M, De Smet I, Beeckman T. 2013. The CEP family in land plants, evolutionary analyses, expression studies, and role inArabidopsisshoot development.Journal of Experimental Botany, 64, 5371–5381.
Roberts I, Smith S, Stes E, De Rybel B, Staes A, Van De Cotte B, Njo M F, Dedeyne L, Demol H, Lavenus J, Audenaert D,Gevaert K, Beeckman T, Smet L D. 2016. CEP5 and XIP1/CEPR1 regulate lateral root initiation inArabidopsis.Journal of Experimental Botany, 67, 4889–4899.
Song W, Liu L, Wang J, Wu Z, Zhang H, Tang J, Lin G, Wang Y, Wen X, Li W, Han Z, Guo H, Chai J. 2016. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth.Cell Research, 26,674–685.
Su H, Zhang S, Yuan X, Chen C, Wang X F, Hao Y J. 2013.Genome-wide analysis and identification of stressresponsive genes of the NAM-ATAF1, 2-CUC2 transcription factor family in apple.Plant Physiology and Biochemistry,71, 11–21.
Sui Z, Wang T, Li H, Zhang M, Li Y, Xu R, Xing G, Ni Z, Xin M. 2016. Overexpression of peptide-encodingOsCEP6.1results in pleiotropic effects on growth in rice (O. sativa).Frontiers in Plant Science, 7, 228.
Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H,Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling.Science, 346, 343–346.
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A,Kalyanaraman A, Fontana P, Bhatnagar S K, Troggio M,Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V,et al. 2010. The genome of the domesticated apple(Malus×domesticaBorkh.).Nature Genetics, 42, 833–839.
Wang J, Guo M Z, Xing L L. 2012. FastJoin, an improved neighbor-joining algorithm.Genetics and Molecular Research, 11, 1909–1922.
Wang X, Zhang S, Su L, Liu X, Hao Y. 2013. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family inMalusdomesticawith a functional characterization ofMdLBD11.PLOS ONE, 8, e57044.
Xu J, Xing S, Zhang Z, Chen X, Wang X. 2016. Genome-wide identification and expression analysis of the tubby-like protein family in theMalusdomesticagenome.Frontiers in Plant Science, 7, e25198.
Zhao T, Liang D, Wang P, Liu J, Ma F. 2012. Genome-wide analysis and expression profiling of the DREB transcription factor gene family inMalusunder abiotic stress.Molecular Genetics and Genomics, 287, 423–436.
 Journal of Integrative Agriculture2018年2期
Journal of Integrative Agriculture2018年2期
- Journal of Integrative Agriculture的其它文章
- Experimental infectivity of Theilerialuwenshuni and Theileria uilenbergi in Chinese Kunming mice
- Development of a sensitive and reliable droplet digital PCR assay for the detection of ‘Candidatus Liberibacter asiaticus’
- One size fits all? Contract farming among broiler producers in China
- Designing price-contingent vegetable rotation schedules using agent-based simulation
- The effects of aeration and irrigation regimes on soil CO2 and N2O emissions in a greenhouse tomato production system
- The efficiency of long-term straw return to sequester organic carbon in Northeast China’s cropland
