AssessmentofVariationinMorpho-PhysiologicalTraitsandGeneticDiversityin Relation to Submergence Tolerance of Five Indigenous LowlandRice Landraces
Jijnasa Barik, Vajinder Kumar, Sangram K. Lenka, Debabrata Panda
Research Paper
AssessmentofVariationinMorpho-PhysiologicalTraitsandGeneticDiversityin Relation to Submergence Tolerance of Five Indigenous LowlandRice Landraces
Jijnasa Barik1, Vajinder Kumar2, Sangram K. Lenka3, Debabrata Panda1
()
The present study evaluated submergence responses in 88 lowland indigenous rice (L.) landraces from Koraput, India, to identify submergence-tolerant rice genotypes. In pot experiments, variations in survival rate, shoot elongation, relative growth index, dry matter, chlorophyll, soluble sugar and starch contents were evaluated in two consecutive years under well-drained and completely submerged conditions. Principal component analysis showed that the first three axes contributed 96.820% of the total variation among the landraces, indicating wide variation between genotypes. Major traits such as survival rate, relative growth index, soluble sugar and starch contents appeared to be important determinants of phenotypic diversity among the landraces. Phenotypic coefficient of variance was higher than genotypic coefficient of variance for all the traits and all showed high heritability (90.38%–99.54%). Five rice landraces (Samudrabali, Basnamundi, Gadaba, Surudaka and Dokarakuji) were the most tolerant to submergence. When submerged for up to 14 d, Samudrabali, Basnamundi and Godoba were notable for having greater survival rates than a standard submergence tolerant variety FR13A, and also notable for elongating more vigorously and accumulatingmorebiomass.Thesethreelandracesmaythereforebe especially useful in lowland rice growing areas that are affected by both moderate stagnant water andflash flooding.MoleculargenotypingrevealedthatthesubmergencetoleranceofSamudrabali,BasnamundiandGodobaislinkedto thepresenceofoneormoredifferentlociand itmay wellproveusefulforbreedingimproved submergencetolerantricevarieties,therebyassisingtoimproveyieldstabilityintherainfedlowlandagro-ecosystem.
genetic variability; genotyping; indigenous rice; submergence tolerance;gene
Flooding-inducedsubmergenceisamajorstressthatlimitsrice(L.)productioninrainfedlowland areasofSoutheastAsia. Itisbecomingamoreseriousissuebecausemodernhighyieldingricevarietiesareill-equippedtotoleratetheincreasesinfarmlandfloodingresultingfromclimatechange(Ismail et al, 2013; Singh et al, 2017; Afrin et al, 2018). A total of 22 million hectares of rice-growingareas are subject tounscheduled submergenceannuallytherebythreateningthelivelihoodofmorethan100millionpeople(Sarkaretal,2006;Singhetal,2016).This,togetherwithariseinthehumanpopulation,threatensfood securityintheflood-pronerainfedlowlands (Singh et al, 2017). Breeding for submergence tolerancewilltherefore becrucialformaintainingstableyieldsinrainfedlowlandecosystem(Daretal,2017;Goswamietal,2017).
The geneticpoolofsubmergence-tolerantricecultivarsis very small. Tolerant rice lines such as FR13A,FR43A, Goda, Heenati, Kurkaruppan, Thavalu 15325 and Thavalu 15314 were identified many years ago but few if any have been discovered more recently (Mazerado and Vergara, 1982; Ram et al, 2002; Sarkar et al, 2006; Singh et al, 2017). A major QTL, designated, was identified in FR13A, a traditional landrace from Odisha (Orissa), India (Sarkar et al, 2006; Xu et al, 2006; Rahman and Zhang, 2016).controls most of the submergence tolerance characters (Neeraja et al, 2007; Septiningsih et al, 2009),and has been successfully introgressed into some popular varieties, such as Swarna, Samba Mahsuri, IR64, RC245, RC249, and a number of improved submergence tolerant high-yielding varieties are developed (Sarkar et al, 2009; Bailey-Serres et al, 2010; Singh et al, 2016; Afrin et al, 2018). However, these improved varieties suffer from being short in stature, thus unsuitable for most of the lowland areas where depth and duration of flooding are high (Iftekharuddaula et al, 2016; Goswami et al, 2017). Farmers therefore hesitate to adopt these genotypes because of poor performance under varying water depths (Afrin et al, 2018). Additionally, the degree of tolerance to submergence not only varies with the genetics of the cultivar but also changes with developmental stage and environmental parameters (Colmer and Pedersen, 2008; Panda et al, 2008; Das et al, 2009). More significantly, submergence is a polygenic trait and theQTLdoes not completely represent the trait alone (Mohanty et al, 2000; Septiningsih et al, 2012; Singh et al, 2014). Several secondary QTLs influencing tolerance have also been identified and located on chromosomes 1, 2, 5, 7, 10 and 11(Toojinda et al, 2003; Sarkar et al, 2006; Septiningsih et al, 2012). Therefore, new submergence-tolerance QTLs that complementneed to be confirmed or identified for breeding high-level submergence tolerant rice variety. Thus, identification of new genetic resources that have no impact on plant height is needed to develop adaptable submergence tolerant varieties.
The success of crop improvement, especially for stressful environments, depends on effective phenotyping based on greater understanding of plant performance under stress conditions (Kuanar et al, 2017). The responses of plants to flooding that confer tolerance are varied, involving physiological and molecular changes (Sarkar et al, 2006; Singh et al, 2017) having been extensively studied in the pasttwo decades (Jackson and Ram, 2003; Sarkar et al, 2006; Bailey-Serres and Voesenek, 2008). Important physiological traits associated with submergence tolerance include maintenance of high carbohydrate, minimum underwater elongation, retention of chlorophyll, maintenance of photosynthetic activity and anti-oxidative protection (Ram et al, 2002; Sarkar et al, 2006; Panda et al, 2008). Precise physiological and molecular marker-based assessment provides information about the extent of genetic diversity, which assists in effective breeding programs (Iftekharuddaula et al, 2016). Due to the heterogeneity in flood-prone ecosystem, different types of traditional rice cultivars are being grown by farmers to suit their local circumstances. These local landraces, low yielding but adapted to different types of flooding stress,could be a useful source of genetic variation for QTL mapping, and they could improve the adaptability of rice to flood-prone areas (Sarkar and Bhattacharjee, 2011; Singh et al, 2017).
The Koraput district of Odisha, India, is rich in indigenous rice varieties and one of the centers of origin for Asian cultivated rice (Patra and Dhua, 2003; Roy et al, 2016). Indigenous rice landraces cultivated by traditional farmers in this region may contain a considerable genetic diversity and thus hold promise for hybridization programmes (Patra and Dhua, 2003; Arunachalam et al, 2006). Indigenous landraces of rice are thus considered as a reservoir of many useful genes, the majority of which remain untapped (Samal et al, 2018). There is a dearth of phenotypic knowledge, genetic variability studies and molecular profiling reports of landraces from Koraput with respect to submergence tolerance. The aim of the present study was to access the genetic diversity and genotypic variability of morpho-physiological traits among selected indigenous rice landraces of Koraput and examine any relationships among the genotypes. This will assist to identify varieties more accurately and prosecute breeding programs focused on submergence tolerance.
MATERIALS AND METHODS
Plant materials
Eighty-eight indigenous lowland rice (L.) landracesfromKoraput,India,wereanalyzedalongwitha floodingtolerantvarietyFR13Aanda susceptiblevarietyIR42aschecks.Landraceseedswere collected from the Mankombu Sambasivan Swaminathan Research Foundation (MSSRF), Jeypore, and the tolerant and susceptible check varieties were from Indian Council of Agricultural Research-National Rice Research Institute, Cuttack, India.
Screening of indigenous rice landraces for submergence tolerance
The submergence tolerance screening was performed for two consecutive years (2016 and 2017) in the experimental garden of Central University of Orissa, Koraput, India (82o44′54′′ E and 18o46′47?? N, 880 m above the mean sea level and average rainfall 1500 mm) during the rice-growing season (March to September). Uniform sized seeds of each genotype were selected and kept at (48±2) oC for 5 d to break the dormancy. All the rice genotypes were then directly seeded in earthen pots containing 2 kg of farm soil and farmyard manure in a 3: 1 ratio. Each pot was supplied with 80 mg urea, 192 mg single super phosphate (P2O5), and 70 mg murate of potash (K2O). Plants were regularly irrigated with tap water and subjected to natural solar radiation, with daily maximum photosynthetic photon flux density, air temperature and relative humidity being about (1360±20)μmol/(m2?s), (31.6 ± 2.0)oC and 65%–75%, respectively. Healthy 21-day-old seedlings were completely submerged in concrete screening tank (3.0 m × 3.0 m × 1.3m) present outside under 1m of water for 14 d and then de-submerged and kept in the open air for another 7 d. One set of plants from each landrace was not submerged but watered regularly to serve as controls. Floodwater pH, water temperature and oxygen concentration were measured at 06:00 and 17:00 h with a water analyzer kit (Syland, Heppenheim, Germany) every other day. Light intensity at 60cm water depth or at the vicinity of the underwater canopy level ranged from 209 to 275μmol/(m2?s)and was 1223–1460μmol/(m2?s)above the water surface. The oxygen concentration at the same water depth was 2.4–3.1 mg/Lat 06:00 h and 4.5–4.8 mg/Lat 17:00 h. The temperature was being 30.4oC to 32.3 oC throughout the period of the experiment. There were three replications in each year for each treatment and pooled data were statistically analyzed and presented.
Measurement of morpho-physiological traits
Fresh and dry weights of five different plants in each replication were taken and dry weight was determined after 3 d at 70 oC. Seedling survival ratewasestimatedafter14dofsubmergencetreatmentfollowedbyde-submergencefor7d.Atthesametime,plantheightandshootelongation weremeasured(Dasetal,2009).Relativegrowthindex(RGI)and dry mass (DM) were calculated according to Bhattacharjee (2008) as follows:
RGI (%) = (Dry weight of submerged seedling/ Dry weight of control seedling) ×100
DM (%) = (Dry weight of seedling/ Fresh weight of seedling) ×100
Measurement of chlorophyll content, SPAD index and carbohydrate content
Leafchlorophyllwasmeasuredbytaking100mgfresh leaves in a 25 mL capped-measuring tube containing 10mL of 80% cold acetone. After extractions for 48 h in a refrigerator (4 oC), chlorophyll was measuredspectro-photometricallyasabsorbanceat663and645nmandcalculatedusingtheequationsofArnon(1949).Leaf greennesswasestimatedastheSPADindexusingthefullyexpandedleafoffivedifferentplantsandaSPAD502 chlorophyllmeter(KonicaMinolta Sensing, Osaka, Japan) that measures intensity oflighttransmittedat650nm (Shrethaetal,2012).Soluble sugar and starch concentrations were estimated (threereplications)after14dof submergencetreatment,followingtheprocedureofYoshidaetal(1976).Briefly,foreachmeasurement,shoot samplesoffiveplantswereoven-dried,groundintoafinepowderandextractedusing80%ethanol.The extractwasthenusedforsolublesugaranalysisafteraddition of Anthrone reagent, followed by measurement of absorbanceat630nm.Aftersugarestimation,theresiduewastreatedwith9.2mol/Lperchloricacidandusedforstarch estimations.
Measurement of genetic variability
The genetic variability of different morpho-physiological parameters among the indigenous rice landraces was estimated by calculating phenotypic and genotypic coefficients of variation. The genotypic variance (σ2G) and phenotypicvariance(σ2P)werecalculatedasperSteeletal(1997).Thephenotypiccoefficient of variation (PCV) and genotypic coefficient ofvariation(GCV)werecalculatedaccording to BurtonandDevane(1953).Broad-sense heritability(2)andgeneticadvancewerecomputedaccordingtoJohnsonetal(1955).
Molecular profiling of selected indigenous rice landraces
After physiological screening, the five most submergence tolerant indigenous rice landraces (Samudrabali, Basnamundi, Godoba, Surudaka and Dokrakuji) were selected for molecular genotyping study along with tolerant (FR13A) and susceptible (IR42) check varieties. Genotyping of the selected genotypes was done by taking five reported simple sequence repeat (SSR) markers linked to submergence tolerance QTL from different chromosomes in rice (Angaji et al, 2010; Septiningsih et al, 2013) (Table 3). For this, three markers within thegene, one each within,and, and two markers linked to QTLs for flooding tolerance during germination,and, were used. Detailed sequence information of these markers were collected from Septiningsih et al (2013) and Angaji et al (2010), which are available in the NCBI database (http://www.ncbi.nlm.nih.gov/).
Rice genomic DNA was isolated by a standardized protocol (Dey et al, 2005).Total genomic DNA was extracted and purified from the young leaves by a modified cetyl-trimethyl ammonium bromide (CTAB) method described by Murray and Thompson (1980). Briefly, fresh leaves (100 mg) were collected from 10-day-old seedlings of each genotype. Isolated total DNA was dissolved in 50 μL of 1× TE (Tris-EDTA) buffer and stored in a -20 oC freezer. PCR amplification was carried out by taking 20 μL volumes mixed with 2 μL 10× PCR buffer, 0.3 μL dNTP mixtures (10 mmol/L), 2 μL SSR primer (2 mmol/L), 2 μL genomic DNA (25 ng/μL), 0.2 μLpolymerase (2 U) (Kapa Biosystems, Sigma) and 12 μL ddH2O, following the method given by Panaud et al (1996). The PCR amplification was an initial denaturation at 94 oC for 3 min, followed by 35 cycles of denaturation at 94 oC for 20 s, annealing at 60 oC for 30 s, extension at 72 oC for 45 s and a final extension of 10 min at 72 oC. The amplified products were resolved through 3.0% ethidium bromide stained (1 μg/mL) agarose gel and documented using a gel documentation system (Alpha Imager, Innotech, USA). The different allelic forms (variation in molecular weight of the amplicons) of individual SSR loci were scored for their presence or absence, respectively across the studied rice genotypes. Polymorphic information content (PIC) for each SSR marker was calculated using the following formula (Hwang et al, 2009).
PIC= 1 ? Σ2ij, where= 1 toandPis the frequency ofth allele for theth band scored for a particular marker.
Marker-based population genetics study including the effective number of alleles () (Kimura and Crow, 1964), and’s heterozygosity () (Nei, 1973) was performed using genetic diversity analysis software POPGENE 1.31 (Yeh and Boyle, 1997).
Statistical analysis
All morpho-physiological parameters were analyzed by two-way analysis of variance with the variety and treatment level (control and submerged condition) as main factors. Differences between various parameters were compared by ANOVA using CROPSTAT (International Rice Research Institute, the Philippines) software. The standard deviations (SD) and regression analysis were done in Microsoft Excel 2007. The Duncan’s multiple range test (DMRT) was also applied to test the variation between genotypes for all the studied parameters. Principal component analysis (PCA) and cluster analysis were carried out using different morpho-physiological traits and PAST-3 (Palaeontological Statistics) software.
RESULTS
Morpho-physiological response of studied rice landraces under submergence
Significant (<0.01) variation of survival rate was observed among the indigenous rice landraces after 14 d of submergence (Supplemental Table 1) with survival rate varying from 5.0% to 98.0%. Five landraces (Samudrabali, Basnamundi, Godoba, Surudaka and Dokrakuji) along with the tolerant check (FR13A) variety showed survival rate of more than 92.0% and they were therefore grouped as submergence tolerant. Samudrabali, Basnamundi and Godoba were especially tolerant, showing higher survival rate (97.0%–98.0%) than the tolerant check FR13A. Ten landraces showedsurvival rateof 71.0%to 89.0%andtenintherangeof41.0%–70.0%.Thesewerecategorisedasmoderatelytolerantand moderatelysusceptible to submergence, respectively. Sixty three landraces along with the susceptible check variety IR42 showed poor survival rate (5.0% to 40.0%) and they were categorised as susceptible to submergence. Similarly, seedling growth in terms of RGI was significantly smaller under submergence, and marked varietal differences (<0.01) were observed (Supplemental Table 1). The range of RGI varied from 33.9% to 95.9% under submergence among the genotypes. In particular,certainlandracesandtolerantFR13A exhibited a higher RGI compared to IR42 (Supplemental Table 1). In contrast, shoot elongation was significantly increased under submergence compared to the control plants, varyingfrom 10.5% to 97.4% amongst genotypes (Supplemental Table 1). In particular, the least amount of shoot elongation was recorded in the submergence tolerant FR13A compared to the susceptible IR42.
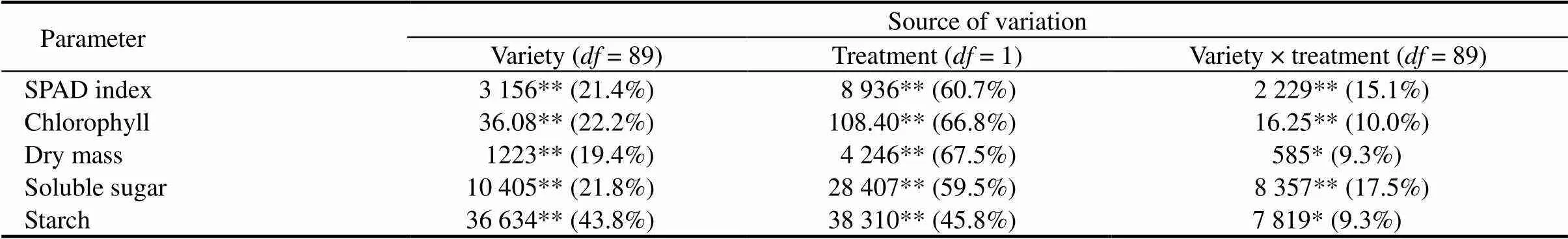
Table 1. Analysis of variance of studied parameters in rice seedlings subjected to submergence.
, Degree of freedom; SPAD, Soil and plant analyzer development. *,< 0.05. **,< 0.01.
DM was significantly (<0.01) inhibited by submergence and a significant (<0.01) varietal difference was observed (Supplemental Table 2). TherangeofDMvariedfrom5.34% to 18.60%after14dof submergence. In particular, the susceptible variety (IR42) exhibited a sharp reduction in DM under submergence compared with the control, whereas some of the indigenous landraces and the submergence tolerant check (FR13A) exhibited higher DM compared to IR42. This parameter was greatly affected by treatment, which accounted for 67.5% of the total variance, followed by 19.4% variance attributable to variety and 9.3% to variety × treatment interaction (Table 1). A similar pattern was also observed for leaf chlorophyll content and SPAD index in tested rice landraces under submergence (Supplemental Table 2). Among the tested landraces, chlorophyll content and SPAD index ranged from 0.14 to 2.00 mg/gand from 3.4 to 31.1, respectively, after submergence. For these parameters, the main causes of variance were the treatment, followed by variety and variety × treatment interaction (Table 1). In addition, there were considerable variations in soluble sugar and starch concentrations among the landraces. Soluble sugar and starch concentrations varied from 8.2–46.2 mg/gand 20.3–74.2 mg/g, respectively. Certain landraces and submergence-tolerant FR13A contained more soluble sugar and starch than submergence-susceptibleIR42. Soluble sugar and starch concentrations were greatly affected by treatment, which accounted for 59.5% and 45.8% of the total variances among the genotypes, respectively (Table 1).
Relationship of different morpho-physiological traits under submergence
Simple regression analysis was performed between morpho-physiological traits and survival rate (%) of landraces under submergence (Fig. 1). Theresultsrevealedstatisticallysignificantpositiveregressions(<0.01)betweensurvival rate andRGI,sugar,starchandchlorophyllcontent.Incontrast,asignificantnegativeregressionwasobservedbetween survival rate and shoot elongation (Fig. 1).

Fig. 1. Relationship of different morpho-physiological traits with survival rate of studied landraces after 14 d of submergence.
Genetic variability analysis among rice landraces
The level of genetic variability, heritability and genetic advance with respect to various morpho-physiological traits in indigenous rice landraces are shown in Table 2. A wide range of variations was observed among the rice genotypes. PCV was slightly higher than GCV for all the traits and low differences were observed between the two. High PCV and GCV values were recorded for survival rate, shoot elongation, chlorophyll, starch and RGI (Table 2). A high heritability estimate was observed in all the traits, ranging from 90.38% to 99.54% (Table 2). Genetic advance as the percentage of means (GAM) for the traits studied ranged from 35.0% to 123.1% (Table 2). High GAM along with high heritability was observed for survival rate, shoot elongation, chlorophyll and starch content.
Principal component analysis and cluster analysis of studied parameters
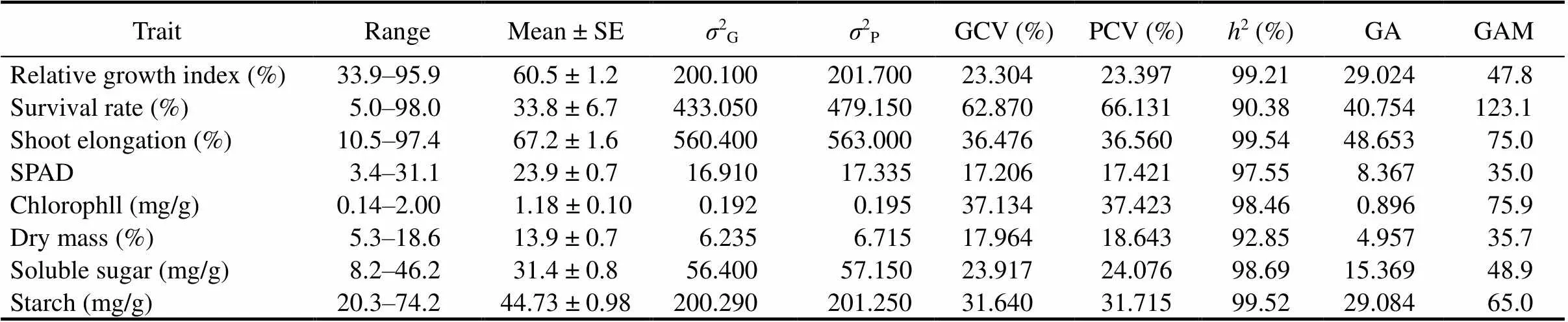
Table 2. Genetic variability parameters for different traits of indigenous rice landraces from Koraput, India.
SE, Standard error;2G, Genotypic variation;2P, Phenotypic variation; GCV, Genotypic coefficient of variation; PCV, Phenotypic coefficient of variation;2, Heritability in broad sense; GA, Genetic advance; GAM, Genetic advance as percentage of the mean.
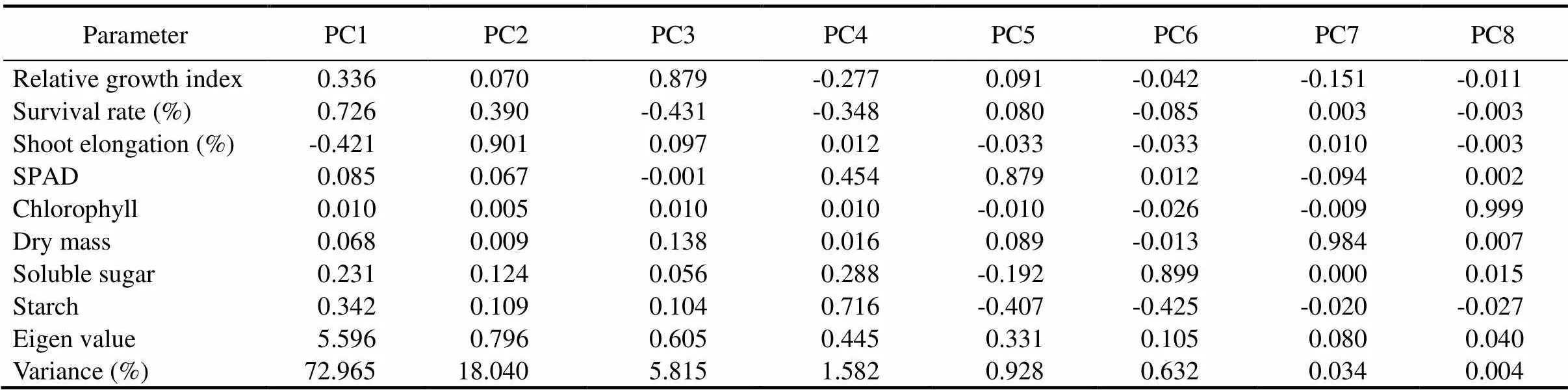
Table 3. Correlations between initial variables with principal component and component loading.
Thegenotypicvariationofdifferentmorpho-physiologicaltraitsundersubmergenceamongthericelandraceswas examinedbymultivariateanalysisincludingPCAandclusteranalysis.Thefirstthree axesofPCAcaptured96.820%ofthetotalvariation.Thissuggestsawidevariability amongthestudiedlandraces(Table 3).PC1,withaneigenvalueof 5.596,accountedfor72.965%ofthevariation amongtheparametersmeasured.InPC1,survival rateexhibitedthehighestpositiveloadingfollowed by RGI, soluble sugar and starch, whereas, in PC2, shoot elongation was constituted mainly of positive effects. Based on the result, survival rate, shoot elongation, RGI, soluble sugar and starch were the major determinants of phenotypic diversity. A scatter plot was drawn between PC1 and PC2. This gave a clear pattern when grouping the genotypes into the factor plane. The first two components of the PCA scatter plot separated the landraces into four quarters with clear separation of submergence tolerantFR13Aand susceptible IR42 (Fig. 2). The indigenous landraces Samudrabali, Basnamundi, Godoba, Surudaka and Dokarakuji along with submergence tolerant FR13A clearly separated from the other landraces and belonged to the most divergent variety present in one quarter. These landraces effectively separated from the other genotypes on the basis of parameters such as survival rate, RGI, soluble sugar and starch.
The unweighted pair group method with arithmetic means (UPGMA) was used to reveal the percent of similarity in different morpho-physiological traits among the rice landraces. Based on the Bray-Curtis paired linkage, the landraces were classified into three major clusters with maximum number (74) landraces included in cluster I with the highest similarity with submergence susceptible variety IR42 (Fig. 3). Thirteen rice landraces along with the tolerant check FR13A were in cluster II. Five tolerant rice landraces Samudrabali, Basnamundi, Godoba, SurudakaandDokarakujiappearedinonesub-clusterandshowedmorethan92%similaritywiththetolerantcheck FR13A. In contrast, eight moderately tolerant genotypes were grouped in a separate cluster having 85% similarity with FR13A. Only one genotype, Kandulakathi, was presented in a separatecluster.
Molecular genotyping of selected indigenous rice landraces
, Number of alleles;, Number of effective alleles;, Expected homozygosity;, Expected heterozygosity;’s, Genetic diversity; PIC, Polymorphism information content.
ThegenotypingresultsobtainedbyanalysingfiveSSRprimerslinkedtosubmergencetoleranceQTLinriceare presentedinTable 4.Tenalleleswereidentifiedamongthegenotypeswithanaverageoftwoallelesperlocus. AmongtheSSRs,Sub1-BC2showedthehighestrangeofallelesize(230–260bp).Thenumbersofeffective allelesrangedfrom1.3to2.0 among the genotypes. The expected homozygosity ()andheterozygosity() rangedfrom0.473to0.736and0.264to0.528,respectively.The’sgeneticdiversityrangedfrom0.245to0.490 among the primers. The level of polymorphism among the seven genotypes was evaluated by calculating the PIC for each of the five SSRs. The highest PICvalue was obtained in primer RM3475 (0.857) followed by Sub1-A203 (0.833), Sub1-BC2 (0.795) and RM478 (0.735).
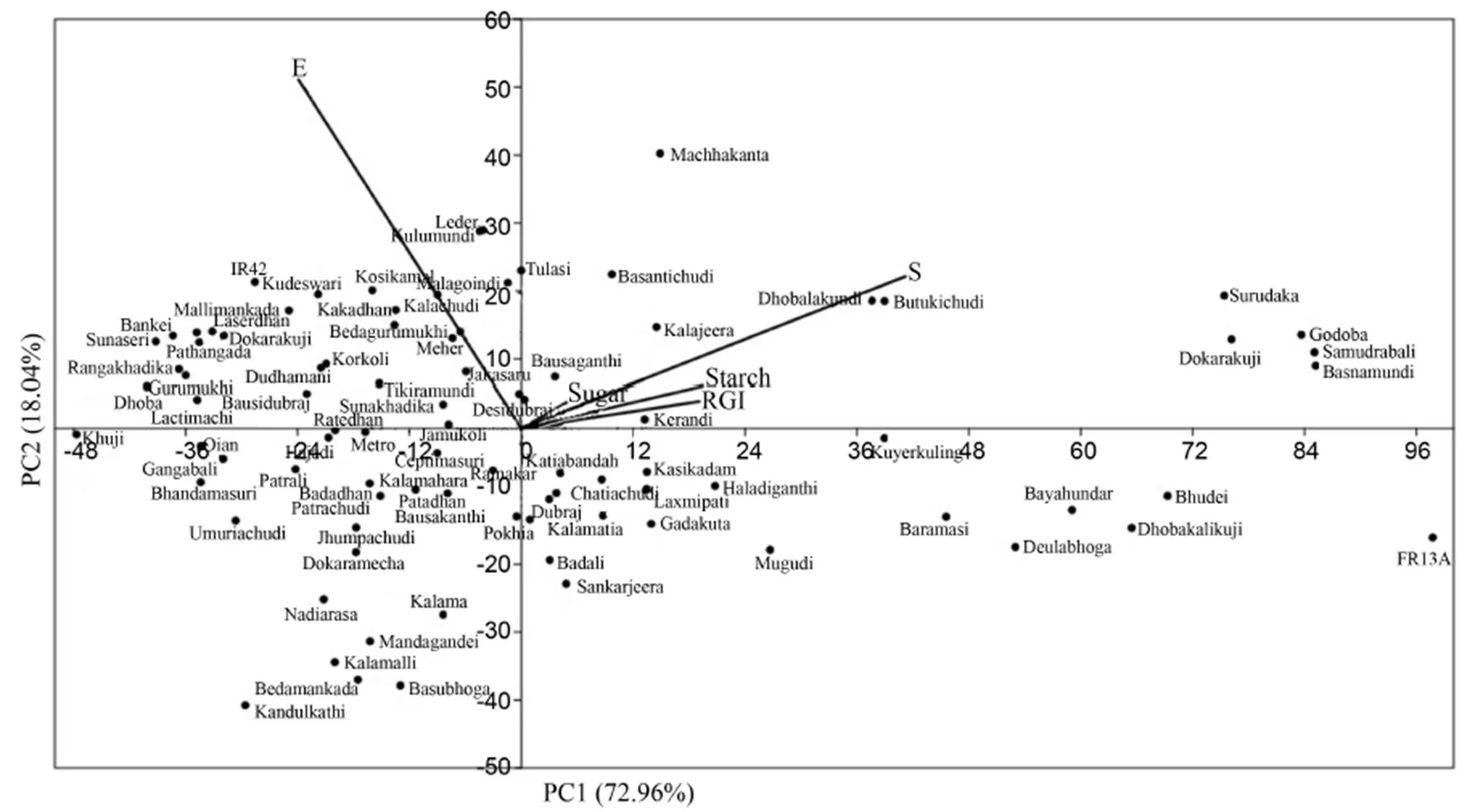
Fig. 2. Principal component analysis (PCA) of different rice landraces showing genotypic relationship in a graphical representation scatter plot on the basis of different morpho-physiological traits under submergence.
The genetic distance among the studied genotypes was calculated with five SSR markers, and it ranged from 0.111 to 0.833 with the highest genetic distance in Surudaka and Dokrakuji(Table 5). The traditional landraces such as Samudrabali, Godoba and Basnamundi showed higher genetic distance againstsubmergence tolerant FR13A compared with the other genotypes, whereas Surudaka and Dokrakuji showed higher genetic distance against submergence susceptible IR42 (Table 5). Cluster analysis based on Jaccard’s similarity paired linkage revealed the percent of similarity in SSR marker data among studied rice genotypes (Fig. 4). The resulting dendrogram showed that similarity forms two major clusters. The indigenous rice landraces Samudrabali, Godoba and Basnamundi along with FR13A were in one cluster having more than 54% similarity whereas Surudaka and Dokrakuji landraces were very close to submergence susceptible IR42.
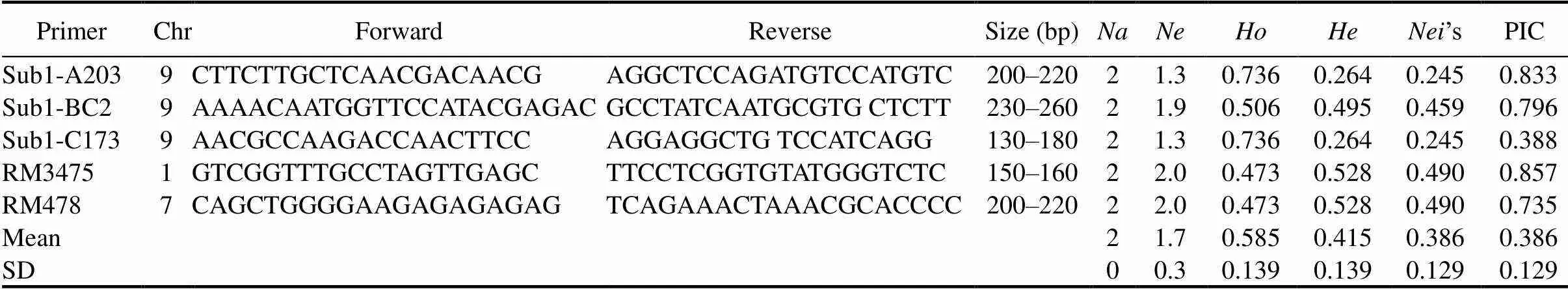
Table 4. Details of molecular marker used for genotyping study and genetic diversity parameters.
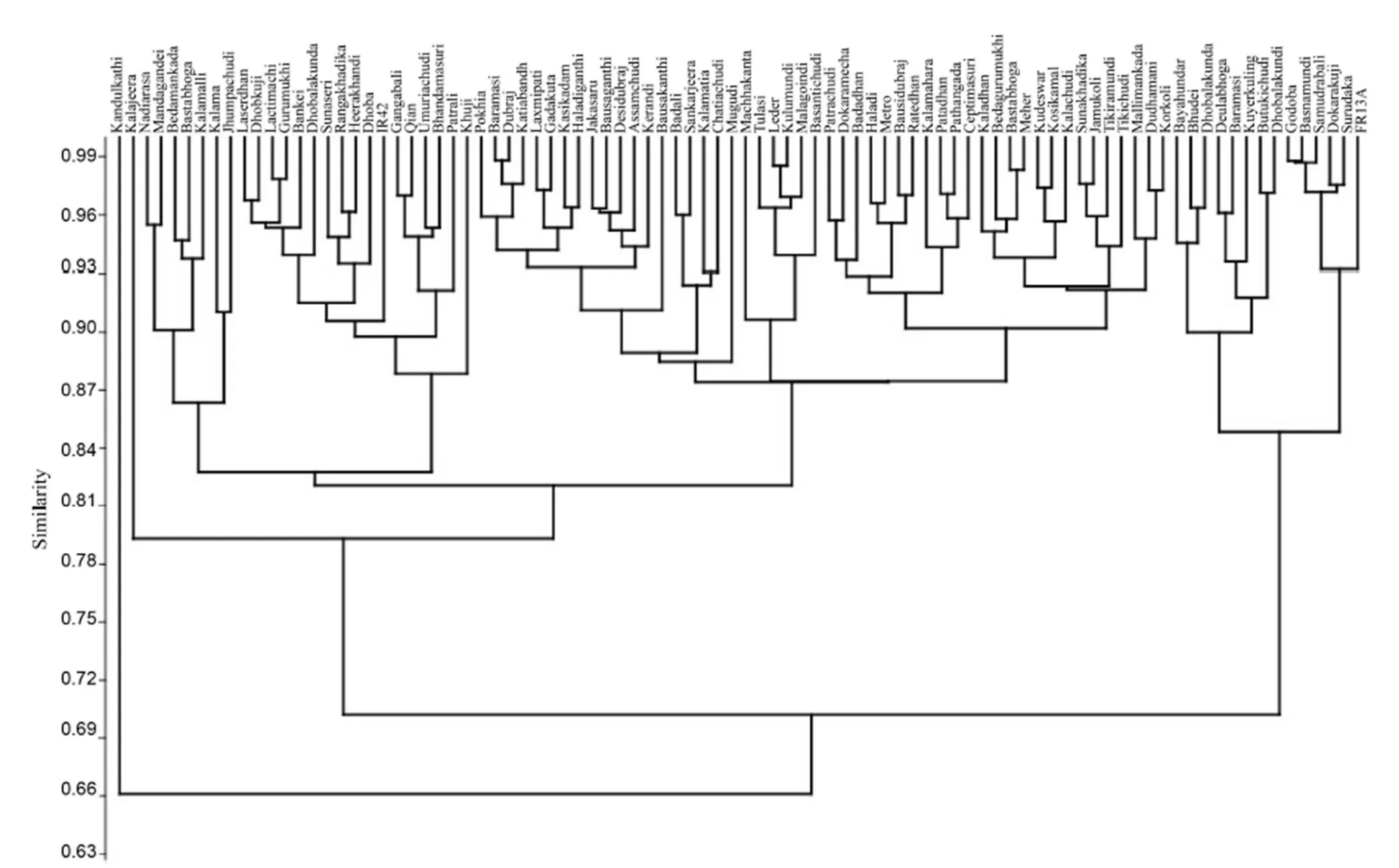
Fig. 3. Similarity index showing in dendrogram of different indigenous rice landraces constructed based on morpho-physiological traits under submergence.
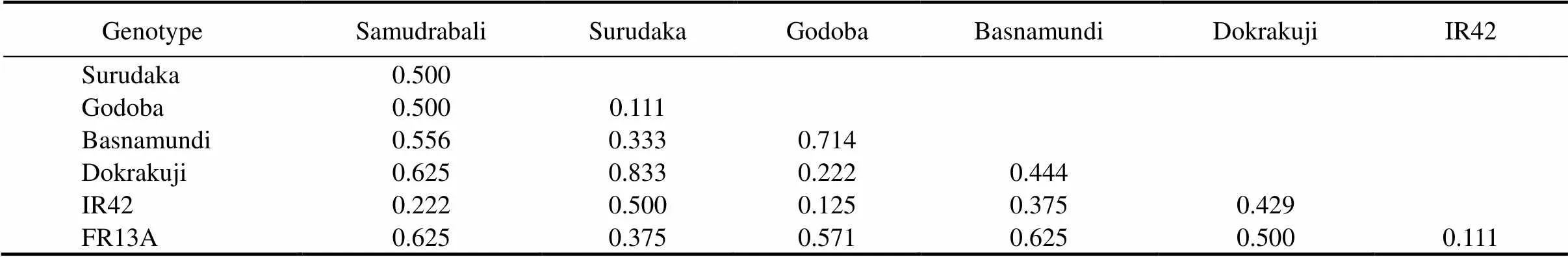
Table 5. Genetic distance between the studied rice genotypes on the basis of SSR markers.
DISCUSSION
Developmentofhigh-yieldingsubmergence-tolerantricevarietiesisalong-cherishedobjectiveofplantbreeders. Forthis,identificationofspecificgenotypesthroughprecisephysiologicalandmolecularmarker-basedassessment provides information aboutthe extentof genetic diversity. This is beneficial when devising effective breeding programs.Duetotheheterogeneityofrainfedlowlandecosystem,manyindigenousricelandracescultivatedby farmersmayserve as potential genetic resources for breeding programs (Singh et al, 2017). In this study, we reported detailed morpho-physiological responses and molecular marker-based assessment of selected lowland indigenous rice landraces from Koraput, India in relation to submergence tolerance. Different rice landraces showed distinct responses to submergence in terms of survival rate, shoot elongation and dry matter accumulation (Supplemental Tables 1 and 2). Submergence significantly impacted on rice seedling survival, and significant varietal differences in the survival rate were observed among the studied landraces. These different responses demonstrated marked differences in sensitivity to submergence among these genotypes. Submergence tolerance is a complex trait which is influenced by the interaction between many traits and environmental conditions (Jackson and Ram, 2003; Das et al, 2009), including the extent of shoot elongation. Submergence significantly enhanced shoot elongation in all the indigenous landraces we examined with the tolerant check variety (FR13A) showing the least elongation compared to all the other genotypes (Supplemental Table 1). RGI and dry matter were also affected by submergence and significant varietal differences were observed among the landraces. Chlorophyll degradation and carbohydrate (sugar and starch) consumption are also more pronounced under submergence in all the studied landraces. In particular, degradations of chlorophyll and carbohydrate content were higher in the susceptible check variety (IR42) compared to the other genotypes (Supplemental Table 2). In this study, survival rate of rice seedlings under submergence was positively influenced by the maintenance of sugar, starch, chlorophyll and plant biomass, whereas it was negatively influenced by shoot elongation. These results are consistent with the earlier observation that lower underwater elongation is beneficial for survival because it lessens the loss of energy reserves while underwater and reduces lodging once water levels recede (Das et al, 2005; Sarkar et al, 2006, 2009; Panda et al, 2008; Singh et al, 2017). In addition, maintaining a higher carbohydrate content and greater biomass during submergence appears crucial for tolerance and helpful for regeneration after submergence (Panda et al, 2008). Maintenance of higher chlorophyll content may also be essential for survival since degradation of chlorophyll decreases photosynthesis (Panda et al, 2006; Panda et al, 2008). Several reports have indicated that better growth under stress conditions is a useful trait when selecting germplasm to improve grain yield (Sarkar and Bhatacharjee, 2011).
Based on the genetic variability analysis, a wide range of variation was observed for the studied parameters among the genotypes. Traits showing a wide range of variation provide ample scope for efficient selection in crop improvement (Mohapatra et al, 2007). In our study, PCV was higher than GCV for all the traits (Table 2). It indicated a high contribution of genotypic effect towards phenotypic expression and these characters were least influenced by the environment. Among the traits, high GAM along with high heritability was observed in survival rate (%), shoot elongation (%), chlorophyll and starch content suggesting that these characters are controlled by additive effectsofgenesandlessinfluencedbyenvironment (Afrinetal,2018).Hence,thesetraitscould,withadvantage,begiventoppriorityduringselectionincrop improvement programs.
PCAmeasurestheimportanceandcontributionofeachcomponenttothe genotypicvariationamongstlandraces(SinhaandMishra,2013).Basedonourresults,thefirstthreeaxesofPCAcapturing96.7%ofthetotalvariationsuggestthewidevariabilityamong the landraces (Table 3). According to Guei et al (2005),thefirstthreeprincipalcomponentsareoftenthemostimportantinreflectingthe variationpatternamongthelandraces,andthecharactersassociatedwiththesearethemostimportantwhen differentiatingvariouslandraces.Basedonthebi-plotanalysis,fiveindigenousricelandraces,Samudrabali, Basnamundi,Godoba,SurudakaandDokarakujialongwithsubmergencetolerantFR13A,clearlyseparatedfrom the otherlandracesandbelongedtomostdivergentgroupinthestudy.Majortraitssuchassurvival rate,RGI, soluble sugar and starch are the major determinants ofphenotypicdiversityamongthelandraces.
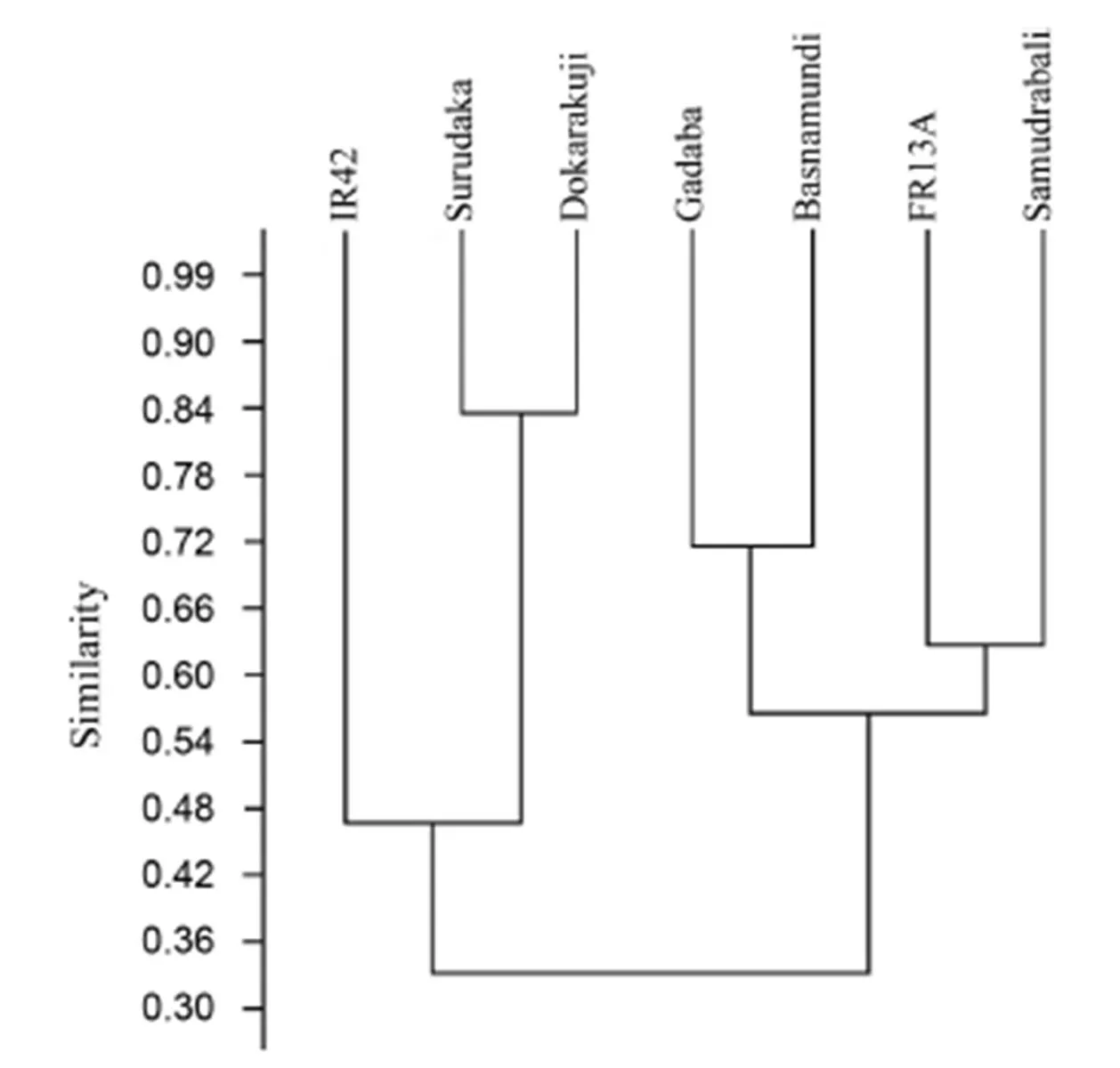
Fig. 4. Dendrogram showing Jackard’s similarity index between rice genotypes based on SSR amplified products.
Based on the morpho-physiological responsesunder submergence,fivericelandracesSamudrabali,Basnamundi,Godoba,SurudakaandDokarakujishowedmorethan 92%similaritywiththetolerantcheckFR13A.Thefindingssuggesttheselandracesarehighlytolerantgenotypes,moststronglyadaptedtosubmergencestress.However,moresignificantly,three landraces Samudrabali, Basnamundi and Godoba showedhighersurvival rateandshootelongationundersubmergencethanthetolerant check FR13A.Itiswidelyrecognisedthatricevarietiesshowinglimitedelongationunderwaterperformbetterafter floodwatersrecedethanvarietiesthatelongatemorequickly(Jackson et al, 1987; Fukao et al, 2011). Thesuppressedelongationarisesbecauseofamutatedgeneinthe QTLthatcheckselongationbyinhibitingthe growth-promotingactionofethylenegastrappedinsidesubmergedplants(Jacksonetal,1987;PandaandSarkar, 2012).Countertothiswell-knownnegativelinkbetweensubmergencetoleranceandshootelongation,wefoundthat Samudrabali,Basnamundi and Godoba elongatedmore than FR13A,but theywerealsomoretoleranttosubmergence. This is more akin to characteristics of deep water rice that involves faster underwater elongation as anescape mechanism(SarkarandBhatacharjee,2011; Goswami et al, 2017). These landraces may be beneficial forlowland ricegrowingareawheresubmergencelastsformorethanafewdaysandthereforemaybesourcesofgenesto generate new genotypes showing superior agronomic performance thanFR13A.
Characterizations of genetic diversity at the DNA level by SSR markers have the potential for estimating the extent of genetic divergence (Hashimoto et al, 2004). Our genotyping results indicated ten alleles among the genotypeswithanaverageoftwoallelesperlocus.Thelevelofpolymorphismwaslowsince only five submergence tolerance-linked markers were used in the study. Thelevelofgeneticdiversityrangedfrom0.244to 0.498amongtheprimers(Table 4).Thislowlevelofgeneticdiversitymaybeduetotheirsimilarorigin,i.e.,all areecotypescollectedfromlowlandareasofKoraput.HigherPICvaluesforprimerRM3475(0.857), Sub1-A203(0.833)andSub1-BC2(0.796)suggeststheirpotentialuseingeneticdiversitystudiesforsubmergence tolerance (Iftekharuddaula et al, 2016).
Based on the allelic diversity of markers,andloci were present in all the five indigenous rice landraces along with FR13A. They were absent inIR42. This finding suggests that submergence tolerance properties in these genotypes may be attributed to the presences ofandloci,which indicates the importance ofandfor differentiating submergence tolerant from submergence susceptible varieties (Septiningsih et al, 2012; Pradhan et al, 2015). In addition, some unique alleles were observed compared to the earlier reported band size ofpoints (Table 4). This is attributed to polygenic submergence traits, which indicates that theQTL does not completely account for submergence tolerance in the landraces we studied (Mohanty et al, 2000; Septiningsih et al, 2012; Singh et al, 2014). The presence of alleles other thanpoints to them as potential sources of new submergence tolerance QTLs and thus of useful genes (Iftekharuddaula et al, 2016). Since some of these landraces showed the highest submergence tolerance capacity in our tests,the information on genetic distance among the landraces can also assist to select tolerance donors in breeding programs (Pradhan et al, 2015).The landraces Samudrabali, Godoba, Basnamundi and submergence tolerant FR13A showed a distinctively higher genetic distance compared with the other genotypes. These highly divergent landraces are therefore promising donors of genes for submergence-tolerance breeding.Finally,clusteranalysisshowedthatSamudrabali,GodobaandBasnamundiformedadistinct sub-cluster along with tolerant FR13A that was clearly separated from the susceptible cultivar IR42.
CONCLUSIONS
Onthebasisofmorpho-physiologicaltraits,widegenotypicvariabilityforsubmergencetolerancewasobserved amongthestudiedlandraces.Majortraitssuchassurvival rate,RGI,soluble sugar andstarchlevelsappearedtobekey determinantsofphenotypicdiversityamongthelandraces. Onthebasisofphysiological screening under submergence, five lowland landraces (Samudrabali, Basnamundi, Godoba, Surudaka andDokrakuji)indigenousto Koraput,India,showedhigherdegreeoftolerancetosubmergence.Amongthesegenotypes, Samudrabali,BasnamundiandGodobaexhibitedthehighestsurvival ratesthatwere unexpectedlyassociated withgreatershootelongationgrowthandbiomassaccumulationundersubmergencecomparedwiththe tolerantcheckFR13A.Theselandracesmaybebeneficialforlowlandricegrowingareathatisaffectedby bothflashfloodingandlongerperiodsofinundation. Molecular genotyping study revealed that these landraces owe their greater submergence tolerance to the presence of one or more loci that are different fromlocus typifying FR13A. These highly genetically divergent landraces maybeuseful fordevelopingnewrice varietieswith high levels of submergencetolerance.
Supplemental DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/ journal/16726308; http://www.ricescience.org.
Supplemental Table 1. Changes of relative growth index, survival rate and shoot elongation in lowland indigenous rice landraces under submergence stress.
Supplemental Table 2. Variations of morpho- physiological parameters in lowland rice landraces of Koraput during seedling stage.
Afrin W, Nafis M H, Hossain M A, Islam M M, Hossain M A. 2018. Responses of rice (L.) genotypes to different levels of submergence., 341(2): 85–96.
Angaji S A, Septiningsih E M, Mackill D J, Ismail A M. 2010. QTLs associated with tolerance of flooding during germination in rice (L.)., 172(2): 159–168.
Arnon D I. 1949. Copper enzymes in isolated chloroplasts, polyphenol oxidase in., 24(1): 1–15.
Arunachalam V, Chaudhury S S, Sarangi S K, Ray T, Mohanty B P, Mishra S. 2006. Rising on Rice: The Story of Jeypore. [MS Thesis]. Chennai, India: Swaminathan Research Foundation.
Bailey-Serres J, Voesenek L A C J. 2008. Flooding stress: Acclimations and genetic diversity.,59: 313–339.
Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D. 2010. Submergence tolerant rice:’s journey from landrace to modern cultivar., 3: 138–147.
Bhattacharjee S. 2008. Calcium-dependent signaling pathway in heatinduced oxidative injury in., 52(1): 137–140.
Burton G W, Devane E H. 1953. Estimating heritability in tall fescue () from replicated clonal material 1., 45(10): 478–481.
Colmer T D, Pedersen O. 2008. Oxygen dynamics in submerged rice ()., 178(2): 326–334.
Dar M H, Chakravorty R, Waza S A, Sharma M, Zaidi N W, Singh A N, Singh U S, Ismail A M. 2017. Transforming rice cultivation in flood prone coastal Odisha to ensure food and economic security.,9(4):711–722.
Das K K, Sarkar R K, Ismail A M. 2005. Elongation ability and non-structural carbohydrate levels in relation to submergence tolerance in rice., 168(1): 131–136.
Das K K, Panda D, Sarkar R K, Reddy J N, Ismail A M. 2009. Submergence tolerance in relation to variable floodwater conditions in rice.,66(3): 425–434.
Dey N, Biswas S, Chaudhuri T, Dey S, De M, Ghose T. 2005. RAPD-based genetic diversity analysis of aromatic rice (L.)., 6(3/4): 133–142.
Fukao T, Yeung E, Bailey-Serres J. 2011. The submergence tolerance regulatormediates crosstalk between submergence and drought tolerance in rice., 23: 412–427.
Goswami S, Kar R K, Paul A, Dey N. 2017. Genetic potentiality of indigenous rice genotypes from Eastern India with reference to submergence tolerance and deepwater traits., 11(12): 23–32.
Guei R G, Sanni K A, Abamu F J, Fawole I. 2005. Genetic diversity of rice (L.).,5: 17–28.
Hashimoto Z, Mori N, Kawamura M, Ishii T, Yoshida S, Ikegami M, Takumi S, Nakamura C. 2004. Genetic diversity and phylogeny of Japanese sake-brewing rice as revealed by AFLP and nuclear and chloroplast SSR markers., 109(8): 1586–1596.
Hwang T Y, Sayama T, Takahashi M, Takada Y, Nakamoto Y, Funatsuki H, Hisano H, Sasamoto S, Sato S, Tabata S, Kono I, Hoshi M, Hanawa M, Yano C, Xia Z J, Harada K, Kitamura K, Ishimoto M. 2009. High-density integrated linkage map based on SSR markers in soybean., 16(4): 213–225.
Iftekharuddaula K M, Ahmed H U, Ghosal S, Amin A, Moni Z R, Ray B P, Barman H N, Siddique M A, Collard B C Y, Septiningsih E M. 2016. Development of early maturing submergence-tolerant rice varieties for Bangladesh., 190: 44–53.
Ismail A M, Singh U S, Singh S, Dar M H, Mackill D J. 2013. The contribution of submergence-tolerant (Sub1) rice varieties to food security in flood-prone rainfed lowland areas in Asia.,152: 83–93.
Jackson M B, Waters I, Setter T, Greenway H. 1987. Injury to rice plants caused by complete submergence:A contribution by ethylene., 38: 1826–1838.
Jackson M B, Ram P C. 2003. Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence., 91: 227–241.
Johnson H W, Robinson H F, Comstock R E. 1955. Estimates of genetic and environmental variability in soybeans.,47(7): 314–318.
Kimura M, Crow J F. 1964. The number of alleles that can be maintained in a finite population.,49(4): 725–738.
Kuanar S R, Ray A, Sethi S K, Chattopadhyay K, Sarkar R K. 2017. Physiological basis of stagnant flooding tolerance in rice., 24(2): 73–84.
Mazaredo A M, Vergara B S. 1982. Physiological differences in rice varieties tolerant and susceptible to completesubmergence.: Proceedings of the 1981 International Deepwater Rice Workshop. Manila, the Phillippines: International Rice Research Institute: 327–341.
Mohanty H K, Mallik S, Grover A. 2000. Prospects of improving flooding tolerance in lowland rice varieties by conventional breeding and genetic engineering., 78(2): 132–137.
Mohapatra M R, Acharya P, Sengupta S. 2007. Variability and association analysis in Okra., 51(1/2): 17–26.
Murray M G, Thompson W F. 1980. Rapid isolation of high molecular weight plant DNA., 8(19): 4321–4325.
Neeraja C N, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard B C, Septiningsih E M, Vergara G, Sanchez D, Xu K, Ismail A M, Mackill D J. 2007. A marker-assisted backcross approach for developing submergence-tolerance rice cultivars., 115(6): 767–776.
Nei M. 1973. Analysis of gene diversity in subdivided populations., 70(12): 3321–3323.
Panaud O, Chen X, McCouch S R. 1996. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (L.).,252(5): 597–607.
Panda D, Rao D N, Sharma S G, Strasser R J, Sarkar R K. 2006. Submergence effects on rice genotypes during seedling stage: Probing of submergence driven changes of photosystem II by chlorophyll a fluorescence induction O-J-I-P transients., 44: 69–75.
Panda D, Sharma S G, Sarkar R K. 2008. Chlorophyll fluorescence parameters, CO2photosynthetic rate and regeneration capacity as a result of complete submergence and subsequent re-emergence in rice (L.)., 88(2): 127–133.
Panda D, Sarkar R K. 2012. Leaf photosynthetic activity and antioxidant defense associated withQTL in rice subjected to submergence and subsequent re-aeration., 19(2): 108–116.
Patra B C, Dhua S R. 2003. Agro-morphoogical diversity scenario in upland rice germplasm of Jeypore tract., 50(8): 825–828.
Pradhan S K, Barik S R, Sahoo J, Pandit E, Nayak D K, Pani D R, Anandan A. 2015. Comparison ofmarkers and their combinations for submergence tolerance and analysis of adaptation strategies of rice in rainfed lowland ecology.,338(10): 650–659.
Rahman B A N M R, Zhang J. 2016. Flood and drought tolerance in rice: Opposite but may coexist.,5(2): 76–88.
Ram P C, Singh B B, Singh A K, Ram P, Singh P N, Singh H P, Boamfa I, Harren F, Santosa E, Jackson M B, Setter T L, Reuss J, Wade L J, Singh V P, Singh R K. 2002. Submergence tolerance in rainfed lowland rice physiological basis and prospects for cultivar improvement through marker-aided breeding.,76: 131–152.
Roy P S, Patnaik A, Rao G J N, Patnaik S S C, Chaudhury S S, Sharma S G. 2016. Participatory and molecular marker assisted pure line selection for refinement of three premium rice landraces of Koraput, India., 41(2): 167–185.
Samal R, Roy P S, Sahoo A, Kar M K, Patra B C, Marndi B C, Gundimeda J N R. 2018. Morphological and molecular dissection of wild rice from eastern India suggests distinct speciation betweenandpopulations., 8: 2773.
Sarkar R K, Reddy J N, Sharma S G, Ismail A M. 2006. Physiological basis of submergence tolerance in rice and implications for crop improvement., 91: 899–906.
Sarkar R K, Panda D, Reddy J N, Patnaik S S C, Mackill D J, Ismail A M. 2009. Performance of submergence tolerant rice () genotypes carrying thequantitative trait locus under stressed and non-stressed natural field conditions.,79(11): 876–883.
Sarkar R K, Bhattacharjee B. 2011. Rice genotype withQTL differ in submergence tolerance, elongation ability during submergence and re-generation growth at re-emergence., 5: 7.
Septiningsih E M, Pamplona A M, Sanchez D L, Neeraja C N, Vergara G V, Heuer S, Ismail A M, Mackill D J. 2009. Development of submergence-tolerant rice cultivars: Thelocus and beyond., 103(2): 151–160.
Septiningsih E M, Sanchez D L, Singh N, Sendon P M D, Pamplona A M, Heuer S, Mackill D J. 2012. Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru., 124(5): 867–874.
Septiningsih E M, Collard B C Y, Heuer S, Bailey-Serres J, Ismail A M, Mackill D J. 2013. Applying genomics tools for breeding submergence tolerance in rice.: Varshney RK, Tuberosa R. Translational Genomics for Crop Breeding. New York, USA: Wiley-Blackwell: 9–30.
Shrestha S, Brueck H, Asch F. 2012. Chlorophyll index, photochemical reflectance index and chlorophyll fluorescence measurements of rice leaves supplied with different N levels., 113: 7–13.
Singh A, Septiningsih E M, Balyan H S, Singh N K, Rai V. 2017. Genetics, physiological mechanisms and breeding of flood-tolerant rice (L.)., 58(2): 185–197.
Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S, Singh N, Prasad K S N, Kondayya K, Rao P V R, Rani M G, Anuradha T, Suraynarayana Y, Sharma P C, Krishnamurthy S L, Sharma S K, Dwivedi J L, Singh A K, Singh P K, Nilanjay, Singh N K, Kumar R, Chetia S K, Ahmad T, Rai M, Perraju P, Pande A, Singh D N, Mandal N P, Reddy J N, Singh O N, Katara J L, Maradi B, Swain P, Sarkar R K, Singh D P, Mohapatra T, Padmawathi G, Ram T, Kathiresan R M, Paramsivam K, Nadarajan S, Thirumeni S, Nagarajan M, Singh A K, Vikram P, Kumwe A, Septiningshih E, Singh U S, Ismail A M, Mackill D, Singh N K. 2016. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network., 242: 278–287.
Singh S, Mackill D J, Ismail A M. 2014. Physiological basis of tolerance to complete submergence in rice involves genetic factors in addition to thegene., 6: plu060.
Sinha A K, Mishra P K. 2013. Morphology based multivariate analysis of phenotypic diversity of landraces of rice (L.) of Bankura district of West Bengal.,9(2): 115–121.
Steel R G, Torrie J H, Dickey D A. 1997.Principles and Procedures of Statistics: A biological Approach. McGraw-Hill.
Toojinda T, Siangliw M, Tragoonrung S, Vanavichit A. 2003. Molecular genetics of submergence tolerance in rice: QTL analysis of key traits., 91(2): 243–253.
Xu K, Xu X, Fukao T, Canalas P, Maghirang-Rodriguez R, Heuer S, Ismail A M, Bailey-Serres J, Ronald P C, Mackill D J. 2006.is an ethylene responsive-factor-like gene that confers submergence tolerance to rice., 442: 705–708.
Yeh F C, Boyle T J B. 1997. Population genetic analysis of codominant and dominant markers and quantitative traits., 129: 157–163.
Yoshida S, Forno D A, Cock J H, Gomez K A. 1976. Laboratory Manual for Physiological Studies of Rice. Manila, the Phillippines:International Rice Research Institute: 14–46.
Copyright ? 2020, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2019.12.004
20 August 2018;
4 December 2018
Debabrata Panda (dpanda80@gmail.com)
(Managing Editor: Fang Hongmin)
- Rice Science的其它文章
- Optimizationof High-Protein Glutinous Rice FlourProduction Using Response Surface Method
- Adoptionand Impactof Modern Rice Varietieson Povertyin Eastern India
- Systematic Characterization of Long Non-Coding RNAs and Their Responses to Drought Stress in Dongxiang Wild Rice
- Differential Expression of Rice Valine-Qlutamine Gene Family in Response to Nitric Oxide and Regulatory Circuit of OsVQ7 and OsWRKY24
- Drought Stress Impairs Grain Yield and Quality of Rice Genotypes by Impaired Photosynthetic Attributes and K Nutrition
- Morpho-Physiological Response of Oryza glaberrima to Gradual Soil Drying

