Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: a systemic review and meta-analysis
Jie-Ying Chen, Ya-Nan Cheng, Lei Han, Feng Wei, Wen-Wen Yu, Xin-Wei Zhang, Shui Cao, Jin-Pu Yu,
1Department of Immunology,2Cancer Molecular Diagnostic Core Laboratory,3Biotherapy Center, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin Key Laboratory of Cancer Immunology and Biotherapy, Tianjin 300060, China
ORIGINaL aRTICLE
Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: a systemic review and meta-analysis
Jie-Ying Chen1, Ya-Nan Cheng2, Lei Han2, Feng Wei1, Wen-Wen Yu1, Xin-Wei Zhang3, Shui Cao3, Jin-Pu Yu1,2
1Department of Immunology,2Cancer Molecular Diagnostic Core Laboratory,3Biotherapy Center, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin Key Laboratory of Cancer Immunology and Biotherapy, Tianjin 300060, China
Objective: A meta-analysis was performed to augment the insufficient data on the impact of mutative EGFR downstream phosphatidylinositol-3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) pathways on the clinical efficiency of epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) treatment of non-small cell lung cancer (NSCLC) patients.
Methods: Network databases were explored in April, 2015. Papers that investigated the clinical outcomes of NSCLC patients treated with EGFR-TKIs according to the status of K-ras and/or PIK3CA gene mutation were included. A quantitative meta-analysis was conducted using standard statistical methods. Odds ratios (ORs) for objective response rate (ORR) and hazard ratios (HRs) for progression-free survival (PFS) and overall survival (OS) were calculated.
Results: Mutation in K-ras significantly predicted poor ORR [OR =0.22; 95% confidence interval (CI), 0.13-0.35], shorter PFS (HR =1.56; 95% CI, 1.27-1.92), and shorter OS (HR =1.59; 95% CI, 1.33-1.91) in NSCLC patients treated with EGFR-TKIs. Mutant PIK3CA significantly predicted shorter OS (HR =1.83; 95% CI, 1.05-3.20), showed poor ORR (OR =0.70; 95% CI, 0.22-2.18), and shorter PFS (HR =1.79; 95% CI, 0.91-3.53) in NSCLC patients treated with EGFR-TKIs. Conclusion: K-ras mutation adversely affected the clinical response and survival of NSCLC patients treated with EGFRTKIs. PIK3CA mutation showed similar trends. In addition to EGFR, adding K-ras and PIK3CA as routine gene biomarkers in clinical genetic analysis is valuable to optimize the effectiveness of EGFR-TKI regimens and identify optimal patients who will benefit from EGFR-TKI treatment.
Non-small cell lung cancer (NSCLC); tyrosine kinase inhibitor (TKI); targeted therapy; K-ras; PIK3CA; meta-analysis
Introduction
Lung cancer remains the leading cause of cancer death in both genders according to the most recent statistics of the American Cancer Society1. Non-small cell lung cancer (NSCLC) accounts for more than 85% of lung cancers2. Most patients present withadvanced NSCLC at the time of diagnosis, and chemotherapy becomes their palliative option. However, the poor improvement in the clinical response and survival outcomes of NSCLC patients who underwent chemotherapy over the last two decades highlights the need for more effective and less toxic treatments3.
Epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) is a small-molecule drug that targets the active adenosine triphosphate binding site of EGFR kinase. Recent studies on patients bearing sensitive EGFR mutation have shown that EGFR-TKIs effectively increase clinical response rate and improve patients’ survival compared with standardchemotherapy, such as cisplatin plus gemcitabine or carboplatin plus paclitaxel, by inhibiting autophosphorylation and activation of downstream signaling pathways4-7. NSCLC patients harboring EGFR mutations benefit more from EGFR-TKI treatment than those without EGFR mutations. However, several studies demonstrated that gene mutations on the EGFR downstream signal pathways are also significant for the response of NSCLC patients to EGFR-TKIs.
EGFR activation elicits its effects via the K-ras/BRAF/mitogenactivated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT/mTOR pathways, which promote tumor proliferation, invasion, migration, and neovascularization8. Mutation in the downstream genes of EGFR signaling pathways may result in receptor-independent pathway activation that renders the tumors unresponsive to EGFR inhibition. K-ras and PI3K are the key regulators on the two aforementioned pathways, respectively. K-ras encodes RAS, a guanosine triphosphate (GTP)-binding protein, which phosphorylates and activates MAPK by interacting with downstream BRAF, leading to a cascade of kinase reactions9. K-ras mutation attenuates the intrinsic GTPase activity of RAS protein, resulting in prolonged RAS activation10. The PIK3CA gene encodes the p110α catalytic subunit of PI3K protein, and its mutation leads to constitutive activation of protein kinase B signaling11. Both pathways play an important role in various cell physiological and pathological processes, such as proliferation, differentiation, apoptosis, and cell migration12-14. Although the corresponding frequencies of K-ras and PIK3CA mutations are approximately 5%-15% and 3%-5%15,16, many studies have reported that K-ras and PIK3CA mutations may have primarily induced resistance to EGFR-TKIs of NSCLC patients17,18.
A previous meta-analysis19indicated a significant correlation between K-ras mutation and clinical response of NSCLC patients treated with EGFR-TKIs. However, the study merely focused on the objective response rate (ORR), and valuable information on the impact of K-ras mutation on the survival of NSCLC patients treated with EGFR-TKIs was not provided because of insufficient data. Similar studies on PIK3CA mutation are rarely reported. Thus, limited information on the clinical significance of gene mutations in the EGFR downstream signal pathways, especially for K-ras and PIK3CA, in NSCLC patients treated with EGFR-TKIs is available.
Therefore, we performed a meta-analysis of published studies to assess the impact of K-ras and PIK3CA mutation on the ORR, progression-free survival (PFS), and overall survival (OS) of NSCLC patients treated with EGFR-TKIs to clarify whether these mutations attenuate the clinical benefits of EGFR-TKI treatment in NSCLC patients.
Materials and methods
Search strategy
We developed a search strategy. An internet search of PubMed, EBSCO, OvidSP, and Wiley Online database was performed in April, 2015. Gefitinib and erlotinib, which are the first-generation EGFR-TKIs, had similar efficacies in NSCLC patients20,21. Thus, a combination of a disease domain (“l(fā)ung cancer”), a treatment domain (“gefitinib”, “erlotinib”, or “EGFR TKI”), and a gene domain (“kras” or “pik3ca”) was used in all fields. The language was limited to English. All retrieved results were sent to EndNote software (EndNote X6, THOMSON REUTERS, US) to automatically and manually check for duplicate studies. After removing the duplicates, the titles and/or abstracts of the remaining results were screened to exclude irrelevant articles. Full texts of relevant articles were obtained and screened further for eligible studies. Bibliographies of relevant articles were handsearched to determine additional eligible studies.
Selection criteria
Two reviewers carefully and independently investigated all studies identified, and consensus was reached after discussion when disagreement in the inclusion or exclusion of studies was encountered. Inclusion criteria were as follows: (I) studies focused on NSCLC patients; (II) studies explored the relation between mutations of K-ras or PIK3CA and outcomes of NSCLC patients treated with EGFR-TKIs; and (III) studies assessed antitumor response using one or more of the following parameters: ORR, PFS, and OS. Distinguishing the predominant effect of the EGFR-TKI treatment was difficult when patients underwent combined therapy treatment. Therefore, exclusion criteria were as follows: (I) patients were not treated with single EGFR-TKIs; and (II) PFS and OS were not calculated from the initiation of EGFR-TKI treatment. When the same patient population was used in several publications, only the most recent, complete, or largest study was included in the meta-analysis.
Data extraction
Data from all eligible studies were extracted independently by two researchers with disagreement settled by discussion. The following data from eligible studies were collected: publication details (such as the first author’s last name, publication year, and country in which the study was performed), trial information (such as inclusion criteria, number of patients assessed, therapy regimens, genes detected and detection methods, and type ofend points used), patient characteristics (such as age, gender, stage, and histology), and outcome measures [such as hazard ratios (HRs) for PFS and OS and their 95% confidence intervals (CIs), log-rank test P values, and ORRs]. PFS and OS were defined as starting from the initial EGFR-TKI treatment. For PFS and OS, the HRs and their 95% CIs were estimated by methods proposed by Tierney et al.22in the absence of published HRs or their 95% CIs. For ORRs, the reported number of objective response (complete response + partial response) and no response (progressive disease + stable disease) in each arm was collected. Quality was assessed independently by two investigators using the Newcastle-Ottawa scale (NOS) for nonrandomized studies (available at http://www.ohri.ca/programs/ clinical_epidemiology/oxford.asp) with consensus on all items through discussion.
Statistical analysis
The relationship between gene mutation and ORR was presented by odds ratio (OR) with 95% CI. The impact of gene mutation on PFS and OS was measured by HR with 95% CI. The pooled ORs were computed for dichotomous variables by the Mantel-Haenszel method, and the pooled HRs and their 95% CIs were estimated by a general variance-based method. Heterogeneity across studies was tested by the χ2-based Q-test and I2statistic. A P value greater than 0.10 for the Q-test and I2statistic with values no more than 50% indicate the lack of heterogeneity among studies. Thus, the fixed-effect model was used for meta-analysis; otherwise, the random-effect model was used. Sensitivity analysis was conducted for meta-analyses by removing one study at a time to test the robustness of the overall results. Potential publication bias was estimated using Begg’s funnel plots and Egger’s linear regression test. All statistical tests were performed with STATA 12.0 (STATA Corporation, College Station, TX). All reported P values were two-sided. Differences were considered statistically significant at P<0.05.
Results
Literature search and study characteristics
The initial search on PubMed, EBSCO, OvidSP, and Wiley Online database in April, 2015 retrieved 2,795 studies. A total of 2,294 articles remained after 501 duplicates were removed. After preliminary screening of titles and/or abstracts, 2,087 non-original or irrelevant studies, 90 book sections, and 74 abstracts or posters of conferences were excluded. Hand search on bibliographies of relevant articles retrieved five additional articles. Thus, full texts of 48 relevant studies were obtained for further investigation. Thirteen articles were further excluded because they were out of scope (12) and they lack relevant data (1). Finally, 19 articles23-41published before 2010, 10 articles17,18,42-49after 2010 and another 6 articles50-55were included. The selection flow diagram is summarized in Figure 1.

Figure 1 Flow diagram of selection process.
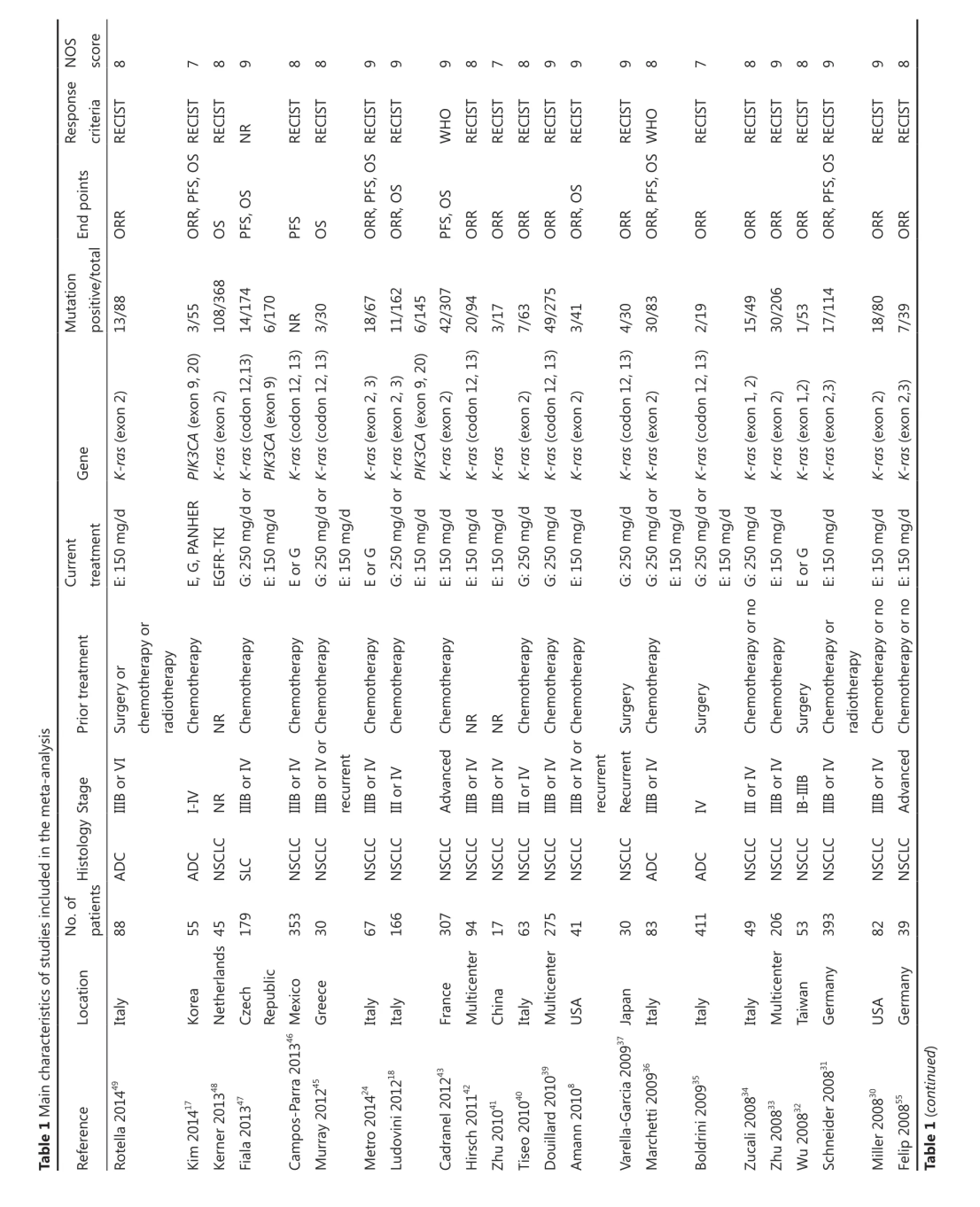
The 35 studies were published from 2006 to 2014. These studies were conducted worldwide: nine from Italy18,26,34-36,40,44,49,53, five from multi-centers (more than two countries or regions)33,39,42,51,52, five from the United States28,30,38,50,54, two from Netherlands29,48, three from Japan23,27,37, two from Korea17,25, two from Germany31,55, and the rest were from Switzerland24, Greece45, France43, Czech Republic47, China41, Mexico46, and Taiwan32. The median age reported in 28 studies ranged from 58 to 75. A total of 3,958 patients were included with a mean sample size of 113 (ranged from 15 to 393). Most studies included patients with NSCLC, with only five studies focused on lung adenocarcinoma17,35,36,49,50, and one focused on lung squamous carcinoma47. Except in two studies17,32, all patients had inoperable stage IIIB or IV or recurrence. Previous treatments included chemotherapy, radiotherapy, surgery, or none. Current treatments of all included studies were monotherapy with EGFR-TKIs. In studies with treatment details, patients were treated with erlotinib or gefitinib according to international standard with one patient who received PF00299804, an irreversible TKI of EGFR, HER2, and HER4, in a study17. Clinical response was evaluated using RECIST criteria56in 31 studies and WHO criteria57in three studies, with one study not reported. Patients with complete or partial responses were classified as responders in all studies. ORR was the end point of 30 studies, PFS in seven studies, and OS in 11 studies. HR and corresponding 95% CI for PFS and OS were calculated from theprimary data reported in the text of one study17, and estimated from the reported summary statistics with method recommended by Tierney in two studies44,47. The quality of all included studies was assessed with NOS. The quality scores of all studies were above 7, with mean score of 8.3.
Biomarker analysis
A total of 33 studies provided the technological details for detecting gene mutations, and 16 studies performed mutation screening using direct sequencing (DS). The rest of the articles included pyrosequencing (1), denaturing capillary electrophoresis (DCE) (3), performance of amplification refractory mutation system (2), polymerase chain reaction (PCR)-restriction fragment length polymorphism (2), mutant-enrich sequencing (ME) (2), and denaturing highperformance liquid chromatography (2). A combination of the aforementioned methods was used in five studies. Mutation in K-ras exons 1, 2, and/or 3 was assessed in 34 studies, and PIK3CA exons 9 and/or 20 in 5 studies. Mutation of EGFR exons 18-21 was detected in all studies.
A total of 573 out of 3,377 evaluable patients were K-rasmutation positive (17.0%), and 18 out of 473 patients were PIK3CA-mutation positive (3.8%). A total of 16 studies reported that K-ras mutation was mutually exclusive with EGFR mutation, and five other studies reported that 10 out of 178 patients positive for K-ras mutation were concomitant with EGFR mutation. Three studies reported that 6 out of 11 patients positive for PIK3CA mutation were concomitant with EGFR mutation. Table 1 shows the main characteristics of studies included in the meta-analysis.
Predictive value of K-ras mutation
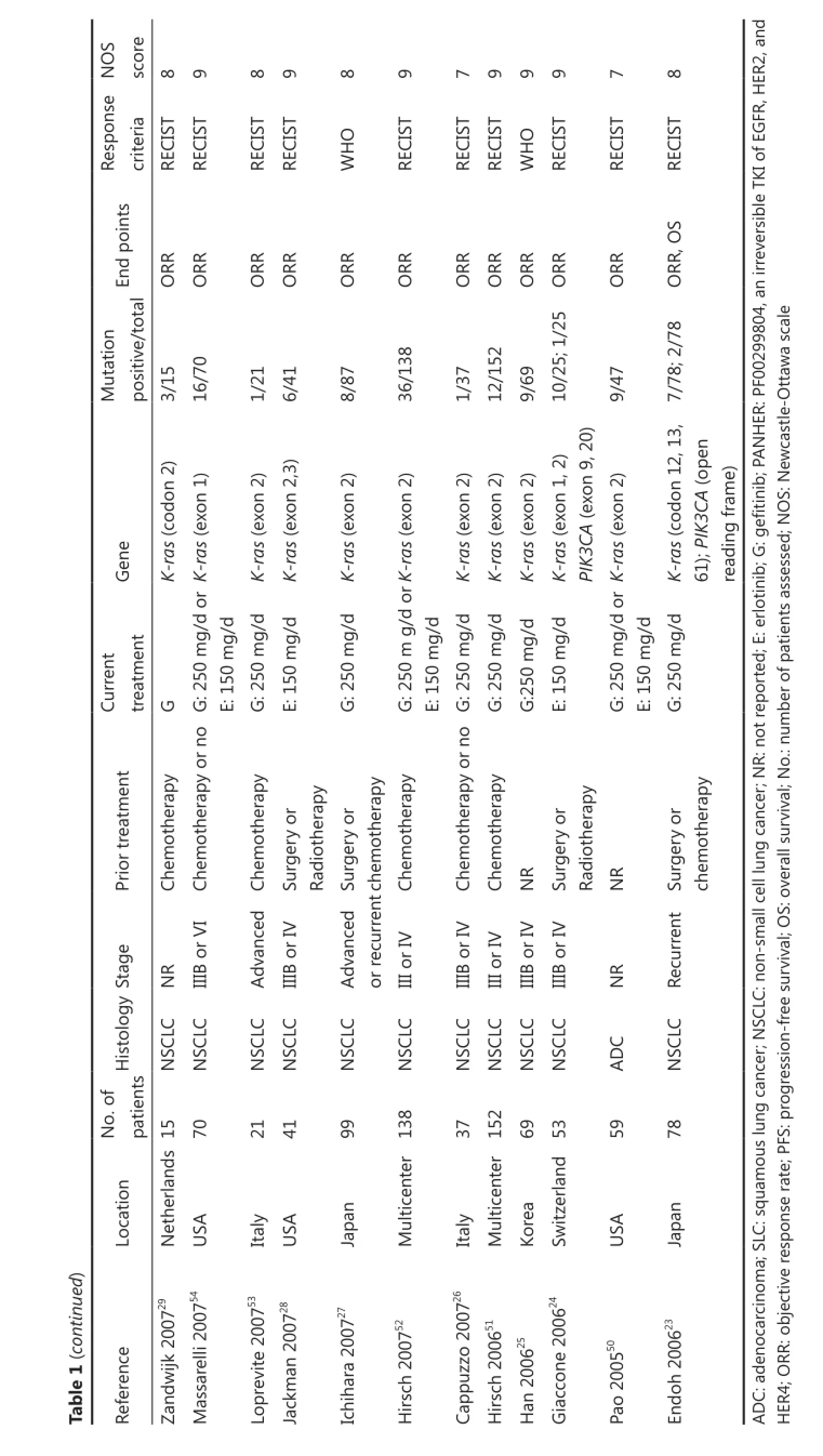
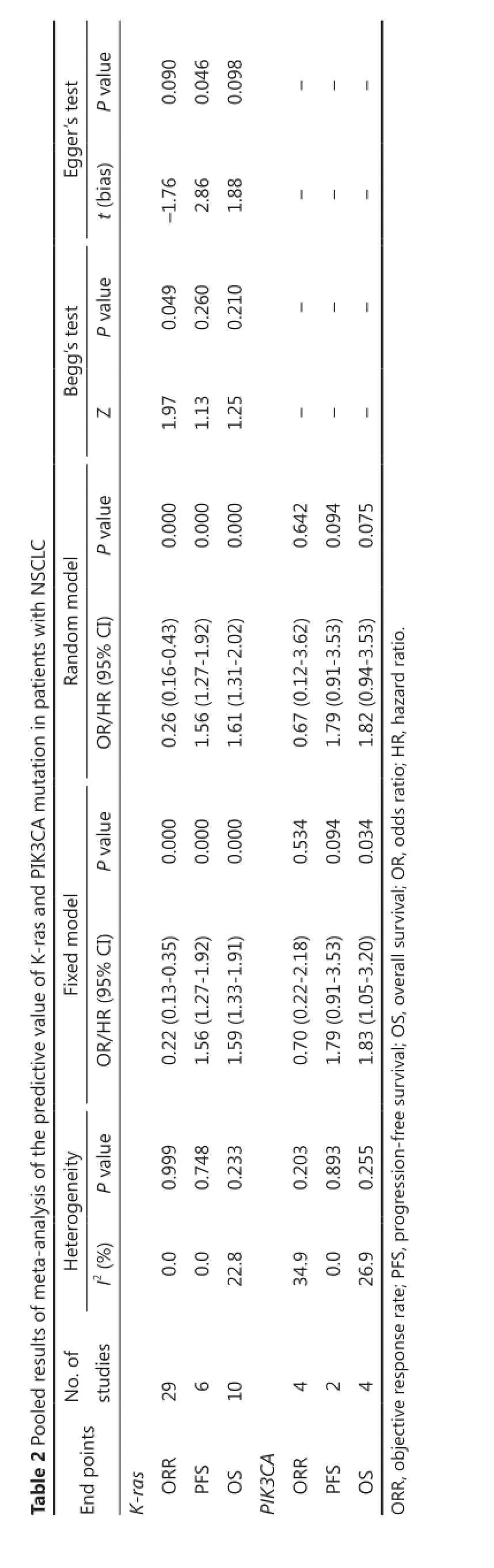
The impact of K-ras mutation on the ORR of NSCLC patients treated with EGFR-TKI therapy was evaluated based on 29 studies (Table 2). K-ras mutation was associated with reduced objective response in NSCLC patients with a pooled OR of 0.22 (95% CI, 0.13-0.35) (Figure 2A). Fixed-effect model was used because heterogeneity across the trials was not significant (I2=0%; P=0.999). The sensitivity analysis indicated that no individual study changed the pooled OR significantly (Figure 2B), suggesting that the result was reliable. Publication bias was significant in Begg’s test (P=0.049), but not in Egger’s test (P=0.090) (Figure 2C). Patients included in two studies39,54apparently originated from the same center. Given that the independence of the two studies could not be confirmed, another analysis excluding the prior one of the aforementioned studies was conducted considering the possibility of duplicate patient population. The pooled OR was 0.22 (95% CI, 0.13-0.35) in a fixed effect model (I2=0%; P=0.998), with publication bias reduced significantly (P values in Egger’s and Begg’s tests were 0.101 and 0.072, respectively).
Data for assessing the impact on PFS according to K-ras mutation status was available in six studies. K-ras mutant patients had shorter PFS compared with wild-type patients with pooled HR of 1.56 (95% CI, 1.27-1.92) (Figure 3A). Fixed-effect model was used when calculating pooled HR for PFS because heterogeneity across trials was not significant (I2=0%; P=0.748). Sensitivity analysis indicated that this result was robust (Figure 3B). Egger’s test revealed slight publication bias (P=0.046), contrary to Begg’s test (P=0.260) (Figure 3C). Thus, a non-parametric“trim-and-fill” method was utilized to adjust the publication bias (Figure 3D). After the trim-and-fill adjustment, two missing studies were added, and the estimated pooled HR was 1.46, with 95% CI ranging from 1.21 to 1.74.
Ten studies were available for analyzing the impact on OS according to K-ras mutation. Results showed that NSCLC patients with K-ras mutation had shorter OS than wild-type patients with pooled HR of 1.59 (95% CI, 1.33-1.91) (Figure 4A). A fixed-effect model was used in calculating pooled HR for OS because heterogeneity across the trials was not significant (I2=22.8%, P=0.233). Sensitivity analysis indicated that the result was stable (Figure 4B). Publication bias was not significant in both Egger’s (P=0.098) and Begg’s tests (P=0.210) (Figure 4C).
To determine the slight heterogeneity across trials in the analysis of the impact of K-ras mutation on the OS of NSCLC patients treated with EGFR-TKIs, we conducted subgroup analysis based on whether K-ras mutation is concomitant with EGFR mutation, previous treatment, and mutation detection method (Table 3). Heterogeneity across trials decreased in most subgroups (Table 3). In addition, a negative effect of K-ras mutation on the OS of NSCLC patients with EGFRTKI treatment was observed in all subgroups, which further confirmed the robustness of the general result.
Predictive value of PIK3CA mutation
S NOscore 8 7 8 9 8 8 9 9 9 8 7 8 9 9 9 8 7 8 9 8 9 9 8 onseO RECISTspO criteriaReRECISTS RECISTNRRECISTRECISTRECISTS RECISTWHRECISTRECISTRECISTRECISTRECISTRECISTRECISTS WHRECISTRECISTRECISTRECISTRECISTS RECISTFS,Ooints S R,PS FS,OFS,OS FS,OR,OR dpEnS ORR R ORR,POSPFS,OPFS OSR R ORORR,OPFS,OORR R,PORORR ORORR R R ORORR,PORORR R OROR/totalORORORtation0 8 7 625 07sitive/84 8 74753 9 06140 Mupo3/558/361013/1146/17NR3/30/618/1116/14/342/9203/177/63/2493/414/30/8302/19/415/2301/53/117/8187/390)) 3)3),22,130),23)3)2,12,13)2,13)2,1n9n9) 2)2,1xo2)2,3n1) 2,1don12,3) ) n9s(exon2)doxon1n1do2)n1do2)n1do2)n1(edo1,2do2)1,2)2,3)2)2,3)CAGene(eK-raPIK3K-raCAs(exons(cos(coK-raPIK3s(coK-raK-ras(exons(exonxoCA(eK-raK-ras(exons(coPIK3K-ras s(exonK-raK-raK-ras(coK-ras(exonK-ras(cos(exonK-raK-ras(cos(exons(exons(exonK-ras(exons(exons(exonororK-raK-raorK-raK-raK-raK-ra/d/dHER mgt or/dmg/d/dor/dmg/d/d/d/d/d/d/dmg/d/d/d/dt en/dmgmg-TKI/d/dmg/dmgmg/dmgmgmgmgmg,PaNrrenmgmgtreatmE:150mg/dmgmgmgmgmgEGG:250CuE:150FRrGE,GG:250EoE:150mgrGEoG:250E:150E:150E:150E:150G:250G:250E:150G:250G:250E:150G:250E:150o G:250E:150rGEoE:150o E:150rno E:150pyornrnotherapyoatmentr r r pyotherapyeryortreotherapyotherapyotherapyotherapyemotherapyotherapyemotherapyemradiotheraotherapyotherapyotherapyery otherapyotherapyery otherapyootherapyootherapyoery emPrioChrgSuchemememememememrgemrgememrgemememlysis NRChChChr ChChChNRNRChChChr SuChSuChChSuChradiotheraChCh-anaI metae orVrreny StagncedrrenrrenncedI-IVIIIBNRorIV IIIBorIV IIIBorIVot IIIBrecuorIV IIIBrIV IIIovaorIV IIIBorIV rIV IIIBorIV IIIoorIVot t IIIBorIV IIIBrIV recuRecuorIV IIIBorIV orIV IIIoadIIIB-IIIB vaIBIIIBIVIIIBaddintheaDaDHistologC CLC C NSSLC CLC CLC CLC NSCLC CLC CLC CLC CLC CLC CLC CLC C C CLC CLC CLC CLC CLC CLC diesincluf tsdeNSNSNSNSNSNSNSNSNSNSaDaDNSNSNSNSNSNS.oNo88patien55s 459 173 3530676 167 304 63754130831 4149063 398239fstur917r2teter253cationerlandCzech blicristicsoy antey thpulticena rmMexicorea lticenGreeceItaly Japan Italy ceanrmKoItaly FranLoNeana inReCh08USItaly Muaractea Italy Italy Italy MuTaiwGeUSGe01) 346 937 001 ched5 lticen449 83348 8 2421Main4 243 039 tinu0 930 0101arra20100936 rcia2015 0101417 5 01nce 1 04420001834 el2043 832000vini20183Fiala201000185er2rray2ble1347 01(con1142Muren2rini2tella2anos-Pli2000832eider20000TaRefeRoKim2rnKeCampMuMetro2doLuCadranHirschu2ZhTiseo2uillard2DoamVarella-GaMarchetti2ldBocaZuu220ZhWuSchnMiller2Felip2ble1Ta
?
?
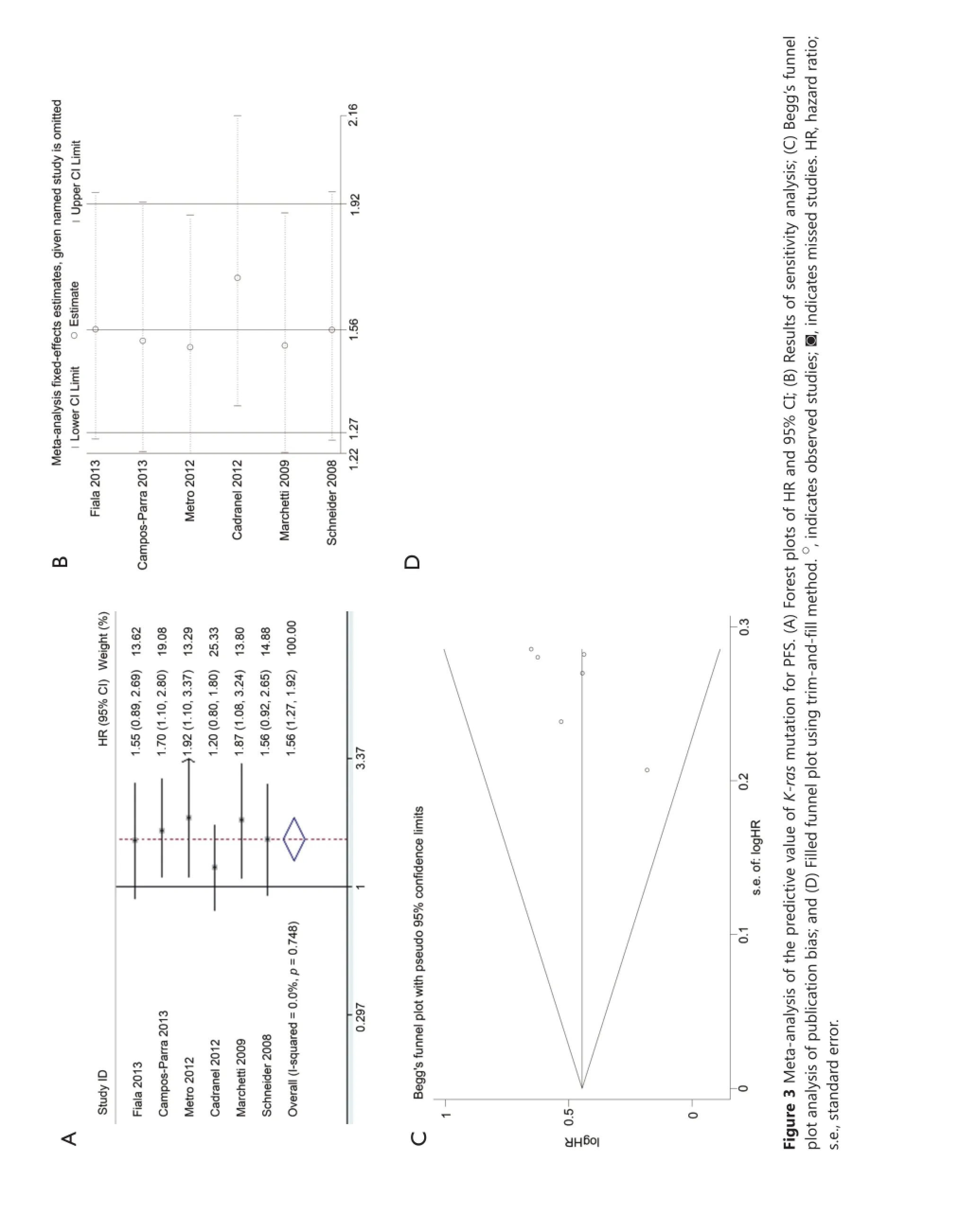
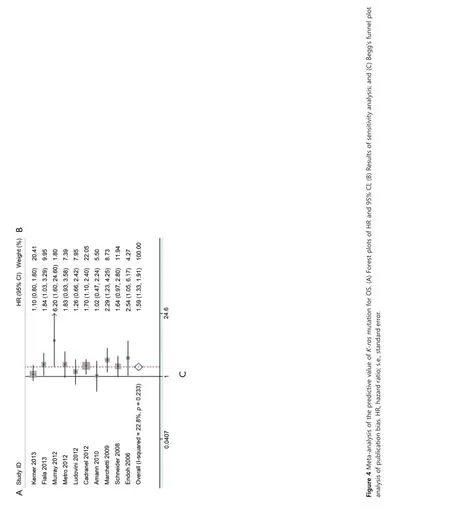
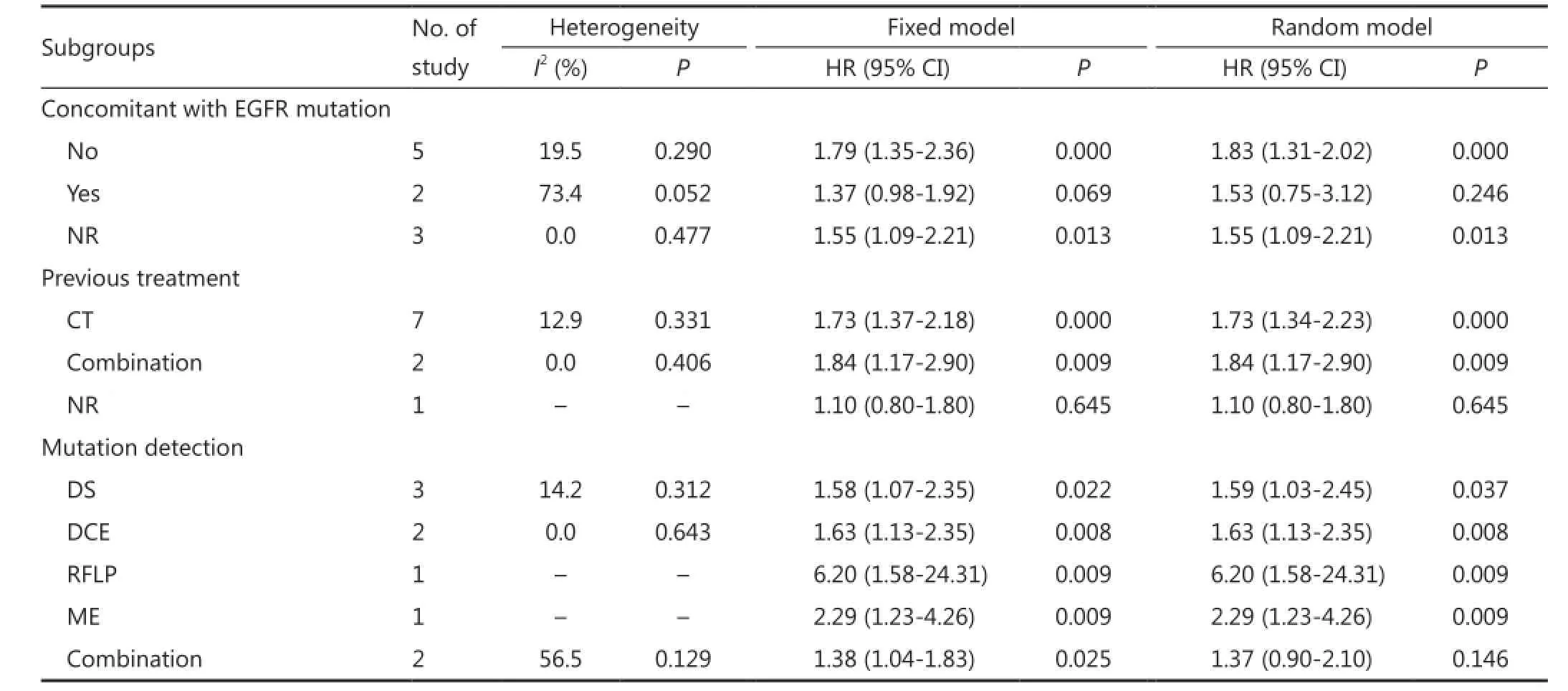
Table 3 Results of subgroup analysis of pooled HRs for OS of patients harboring K-ras mutation with EGFR-TKI treatment
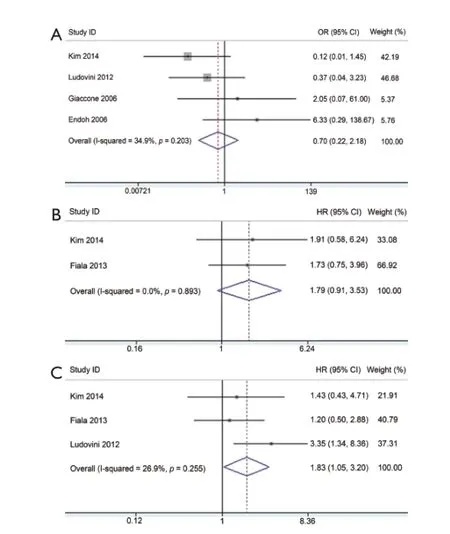
Figure 5 Meta-analysis of the predictive value of PIK3CA mutation. (a) Forest plots of OR and 95% CI for ORR; (B) Forest plots of HR and 95% CI for PFS; and (C) Forest plots of HR and 95% CI for OS. OR, odds ratio; HR, hazard ratio.
Five studies investigated the predictive role of PIK3CA mutation in NSCLC patients (Table 2). Among these, ORR data were available in four studies, PFS data in two studies, and OS data in three studies. PIK3CA mutant NSCLC patients exhibited similar response to EGFR-TKIs compared with wild-type patients with corresponding pooled OR of 0.70 (95% CI, 0.22-2.18) (Figure 5A). Fixed-effect model was used because heterogeneity across studies was not significant (I2=34.9%; P=0.203). The pooled HR of 1.79(95% CI, 0.91-3.53) for PFS in a fixed-effect model (I2=0%; P=0.893) suggested that PIK3CA mutant NSCLC patients had similar PFS compared with wild-type patients when treated with EGFR-TKIs (Figure 5B). However, PIK3CA mutation showed a trend toward a significant adverse effect on OS with a pooled HR of 1.83 (95% CI, 1.05-3.20) in NSCLC patients treated with EGFR-TKIs (Figure 5C). Between-study heterogeneity was not significant; thus, the analysis was performed in the fixed-effect model (I2=26.9%; P=0.255).
Sensitivity analysis and publication bias of all above analyses was not performed because of the relatively limited eligible studies. Subgroup analysis was not conducted because of the relatively small size of included articles.
Discussion
EGFR inhibitor elicits multiple downstream effects, primarily moderated by RAS/RAF/MAPK and PI3K/AKT/mTOR signaling pathways. Rational use of target therapy requires the optimal selection of patients whose tumors are dependent on the activation of these two pathways. The predictive value of gene mutations on these two pathways downstream of EGFR for EGFR-TKI treatment is gradually recognized. This meta-analysis reveals an independent predictive value of K-ras and PIK3CA genetic status on EGFR-TKI therapy.
Coincident with previous report, our results again demonstrated that NSCLC patients harboring K-ras mutation had poor response to EGFR-TKIs. Exclusion of possible duplicate study reduced publication bias significantly and did not alter the pooled result, thus proving the stability of our result. More importantly, we quantitatively demonstrated that such patients had shorter PFS and OS compared with wild-type patients. Given that heterogeneity was zero across the studies in the analysis of the impact of K-ras mutation on PFS of NSCLC patients treated with EGFR-TKIs, a trim-and-fill method was applied to adjust publication bias. The adjusted pooled HR did not alter significantly the primary result, suggesting the dependability of our results. Slight heterogeneity was observed in the meta-analysis of the impact of K-ras mutation on the OS of NSCLC patients treated with EGFR-TKIs. Subgroup analysis showed that if K-ras mutation is concomitant with EGFR mutation, previous treatment and mutation detection method (Table 3) might affect the result. However, a negative effect of K-ras mutation on the OS of NSCLC patients with the EGFRTKI treatment was observed in all subgroups. All these results indicated the adverse impact of mutant K-ras on the response and survival outcomes of NSCLC patients treated with EGFRTKIs. This adverse effect has been proved in other cancers58.
Mutant PIK3CA proteins increase catalytic activity resulting in enhanced downstream signaling and oncogenic transformation in vitro59. Preclinical data showed that introducing activated PIK3CA mutations into EGFR-mutated lung cancer cell lines confers resistance to EGFR-TKIs60. Consistent with this result, our analysis revealed significantly shorter OS, poor ORR, and shorter PFS in PIK3CA mutant NSCLC patients treated with EGFR-TKIs.
The most common mutation of PIK3CA was found in exons 9 and 2061, corresponding to the helical and kinase domains, respectively. The predictive value of PIK3CA as a negative biomarker for anti-EGFR response in colorectal cancer differed in exons 9 and 2062. However, similar study in NSCLC was rarely reported in published articles, and no clear evidence was obtained to show that the impact of mutations in exons 9 and 20 of PIK3CA on anti-EGFR response differ in NSCLC. Thus, further analysis of the predictive value of these two exons separately with enlarged samples size is needed to achieve definite conclusion.
Slight heterogeneity was observed in the analysis of the impact of PIK3CA mutation. Although subgroup analysis could not be conducted because of insufficient data, some diversity on whether PIK3CA mutation was concomitant with EGFR mutation, mutation detecting method, and data extraction method was observed. Coexistence of PIK3CA mutations with EGFR is frequent in lung cancer15,63,64. However, the predictive value of PIK3CA to anti-EGFR treatment in EGFR mutant or wild-type NSCLC is ambiguous at present. The accuracy and specificity of different mutation detection methods also varied, which led to different false positive and false negative rates36. Although extracting time-to-event data according to Tierney was preferable, it failed to circumvent the potential biases associated with relying on published data for meta-analysis as mentioned by the authors. Therefore, despite the slight heterogeneity of the included studies in the analysis of the impact of mutant PIK3CA on the response and survival outcomes of NSCLC patients, our result would be consolidated by increasing sample size.
Despite our efforts to provide an accurate and comprehensive analysis, limitations of our meta-analysis should be addressed. First, most of the included studies were retrospective. Second, not all published studies presented adjusted estimates or had been adjusted by similar potential confounders. Third, limited studies presented PIK3CA mutation data, in which only four studies provided ORR information, two studies provided PFS information, and three studies provided OS information. Thus, increasing sample size of studies will further increase the creditability of adverse effect of PIK3CA mutation on clinical prognosis of NSCLC patients receiving EGFR-TKI treatment.
In conclusion, this meta-analysis indicated that K-ras mutation is probably a valuable predictive biomarker for assessing the clinical response and survival outcomes of NSCLC patients treated with EGFR-TKIs. More importantly, similar trends for PIK3CA mutation were shown in this meta-analysis, although the trends in ORR and PFS were not significant. Increasing sample size of studies will further increase the creditability of adverse effect of PIK3CA mutation on the clinical prognosis of NSCLC patients receiving EGFR-TKI treatment. Mutations of K-ras and EGFR are usually mutually exclusive, and coexistence of mutation in PIK3CA and EGFR is common. Thus, determining the status of K-ras and PIK3CA is valuable to distinguish the optimal patients who will benefit from EGFR-TKI treatment.
Acknowledgements
This work was supported by Key Projects in the National Science & Technology Pillar Program (Grant No. 2013ZX09303001, 2015BAI12B12, and 2015BAI12B15), National Natural Science Foundation of China (Grant No. 81472473 and 81272360), and Tianjin Municipal Commission of Science & Technology Key Research Program (Grant No.13ZCZCSY20300). We thank Dr. Wei-Jia Zhang from the Department of Medicine, Icahn School of Medicine at Mount Sinai for providing constructive suggestions for manuscript preparation.
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29.
2. Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J 2001;18:1059-1068.
3. Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002;346:92-98.
4. Pal SK, Figlin RA, Reckamp K. Targeted therapies for nonsmall cell lung cancer: an evolving landscape. Mol Cancer Ther 2010;9:1931-1944.
5. Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-222.
6. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-246.
7. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-2388.
8. Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160-1174.
9. Thatcher JD. The Ras-MAPK signal transduction pathway. Sci Signal 2010;3:tr1.
10. Gibbs JB, Sigal IS, Poe M, Scolnick EM. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A 1984;81:5704-5708.
11. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554.
12. Spaargaren M, Bischoff JR, McCormick F. Signal transduction by Ras-like GTPases: a potential target for anticancer drugs. Gene Expr 1995;4:345-356.
13. Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell 2000;103:253-262.
14. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011;12:21-35.
15. Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H, et al. Coexistence of EGFR with K-ras, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer 2014;110:2812-2820.
16. Xu J, He J, Yang H, Luo X, Liang Z, Chen J, et al. Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell lung cancer. Cancer Biomark 2011-2012;10:63-69.
17. Kim HR, Cho BC, Shim HS, Lim SM, Kim SK, Chang J, et al. Prediction for response duration to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutated never smoker lung adenocarcinoma. Lung Cancer 2014;83:374-382.
18. Ludovini V, Bianconi F, Pistola L, Pistola V, Chiari R, Colella R, et al. Optimization of patient selection for EGFR-TKIs in advanced non-small cell lung cancer by combined analysis of K-ras, PIK3CA, MET, and non-sensitizing EGFR mutations. Cancer Chemother Pharmacol 2012;69:1289-1299.
19. Mao C, Qiu LX, Liao RY, Du FB, Ding H, Yang WC, et al. K-ras mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer 2010;69:272-278.
20. Burotto M, Manasanch EE, Wilkerson J, Fojo T. Gefitinib and erlotinib in metastatic non-small cell lung cancer: a meta-analysisof toxicity and efficacy of randomized clinical trials. Oncologist 2015;20:400-410.
21. Song Z, Zhang Y. Efficacy of gefitinib or erlotinib in patients with squamous cell lung cancer. Arch Med Sci 2015;11:164-168.
22. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into metaanalysis. Trials 2007;8:16.
23. Endoh H, Yatabe Y, Kosaka T, Kuwano H, Mitsudomi T. PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol 2006;1:629-634.
24. Giaccone G, Gallegos Ruiz M, Le Chevalier T, Thatcher N, Smit E, Rodriguez JA, et al. Erlotinib for frontline treatment of advanced non-small cell lung cancer: a phase II study. Clin Cancer Res 2006;12:6049-6055.
25. Han SW, Kim TY, Jeon YK, Hwang PG, Im SA, Lee KH, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res 2006;12:2538-2544.
26. Cappuzzo F, Ligorio C, Janne PA, Toschi L, Rossi E, Trisolini R, et al. Prospective study of gefitinib in epidermal growth factor receptor fluorescence in situ hybridization-positive/phospho-Akt-positive or never smoker patients with advanced non-small-cell lung cancer: the ONCOBELL trial. J Clin Oncol 2007;25:2248-2255.
27. Ichihara S, Toyooka S, Fujiwara Y, Hotta K, Shigematsu H, Tokumo M, et al. The impact of epidermal growth factor receptor gene status on gefitinib-treated Japanese patients with non-smallcell lung cancer. Int J Cancer 2007;120:1239-1247.
28. Jackman DM, Yeap BY, Lindeman NI, Fidias P, Rabin MS, Temel J, et al. Phase II clinical trial of chemotherapy-naive patients > or = 70 years of age treated with erlotinib for advanced non-small-cell lung cancer. J Clin Oncol 2007;25:760-766.
29. van Zandwijk N, Mathy A, Boerrigter L, Ruijter H, Tielen I, de Jong D, et al. EGFR and K-ras mutations as criteria for treatment with tyrosine kinase inhibitors: retro- and prospective observations in non-small-cell lung cancer. Ann Oncol 2007;18:99-103.
30. Miller VA, Riely GJ, Zakowski MF, Li AR, Patel JD, Heelan RT, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol 2008;26:1472-1478.
31. Schneider CP, Heigener D, Schott-von-Romer K, Gutz S, Laack E, Digel W, et al. Epidermal growth factor receptor-related tumor markers and clinical outcomes with erlotinib in non-small cell lung cancer: an analysis of patients from german centers in the TRUST study. J Thorac Oncol 2008;3:1446-1453.
32. Wu CC, Hsu HY, Liu HP, Chang JW, Chen YT, Hsieh WY, et al. Reversed mutation rates of K-ras and EGFR genes in adenocarcinoma of the lung in Taiwan and their implications. Cancer 2008;113:3199-3208.
33. Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, et al. Role of K-ras and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 2008;26:4268-4275.
34. Zucali PA, Ruiz MG, Giovannetti E, Destro A, Varella-Garcia M, Floor K, et al. Role of cMET expression in non-small-cell lung cancer patients treated with EGFR tyrosine kinase inhibitors. Ann Oncol 2008;19:1605-1612.
35. Boldrini L, Ali G, Gisfredi S, Ursino S, Baldini E, Melfi F, et al. Epidermal growth factor receptor and K-ras mutations in 411 lung adenocarcinoma: a population-based prospective study. Oncol Rep 2009;22:683-691.
36. Marchetti A, Milella M, Felicioni L, Cappuzzo F, Irtelli L, Del Grammastro M, et al. Clinical implications of K-ras mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia 2009;11:1084-1092.
37. Varella-Garcia M, Mitsudomi T, Yatabe Y, Kosaka T, Nakajima E, Xavier AC, et al. EGFR and HER2 genomic gain in recurrent non-small cell lung cancer after surgery: impact on outcome to treatment with gefitinib and association with EGFR and K-ras mutations in a Japanese cohort. J Thorac Oncol 2009;4:318-325.
38. Amann JM, Lee JW, Roder H, Brahmer J, Gonzalez A, Schiller JH, et al. Genetic and proteomic features associated with survival after treatment with erlotinib in first-line therapy of non-small cell lung cancer in Eastern Cooperative Oncology Group 3503. J Thorac Oncol 2010;5:169-178.
39. Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol 2010;28:744-752.
40. Tiseo M, Rossi G, Capelletti M, Sartori G, Spiritelli E, Marchioni A, et al. Predictors of gefitinib outcomes in advanced non-small cell lung cancer (NSCLC): study of a comprehensive panel of molecular markers. Lung Cancer 2010;67:355-360.
41. Zhu YJ, Xia Y, Ren GJ, Wang MZ, Zeng X, Zhang L. Efficacy and clinical/molecular predictors of erlotinib monotherapy for Chinese advanced non-small cell lung cancer. Chin Med J (Engl) 2010;123:3200-3205.
42. Hirsch FR, Kabbinavar F, Eisen T, Martins R, Schnell FM, Dziadziuszko R, et al. A randomized, phase II, biomarkerselected study comparing erlotinib to erlotinib intercalated with chemotherapy in first-line therapy for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3567-3573.
43. Cadranel J, Mauguen A, Faller M, Zalcman G, Buisine MP, WesteelV, et al. Impact of systematic EGFR and K-ras mutation evaluation on progression-free survival and overall survival in patients with advanced non-small-cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project--part 2). J Thorac Oncol 2012;7:1490-1502.
44. Metro G, Chiari R, Duranti S, Siggillino A, Fischer MJ, Giannarelli D, et al. Impact of specific mutant K-ras on clinical outcome of EGFR-TKI-treated advanced non-small cell lung cancer patients with an EGFR wild type genotype. Lung Cancer 2012;78:81-86.
45. Murray S, Karavasilis V, Bobos M, Razis E, Papadopoulos S, Christodoulou C, et al. Molecular predictors of response to tyrosine kinase inhibitors in patients with Non-Small-Cell Lung Cancer. J Exp Clin Cancer Res 2012;31:77.
46. Campos-Parra AD, Zuloaga C, Manríquez ME, Avilés A, Borbolla-Escoboza J, Cardona A, et al. KRAS mutation as the biomarker of response to chemotherapy and EGFR-TKIs in patients with advanced non-small cell lung cancer: clues for its potential use in second-line therapy decision making. Am J Clin Oncol 2013;38:33-40.
47. Fiala O, Pesek M, Finek J, Benesova L, Bortlicek Z, Minarik M. Gene mutations in squamous cell NSCLC: insignificance of EGFR, K-ras and PIK3CA mutations in prediction of EGFR-TKI treatment efficacy. Anticancer Res 2013;33:1705-1711.
48. Kerner GS, Schuuring E, Sietsma J, Hiltermann TJ, Pieterman RM, de Leede GP, et al. Common and rare EGFR and K-ras mutations in a Dutch non-small-cell lung cancer population and their clinical outcome. PLoS One 2013;8:e70346.
49. Rotella V, Fornaro L, Vasile E, Tibaldi C, Boldrini L, Chella A, et al. EGFR and K-Ras mutations in women with lung adenocarcinoma: implications for treatment strategy definition. J Exp Clin Cancer Res 2014;33:77.
50. Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. K-ras mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005; 2: e17.
51. Hirsch FR, Varella-Garcia M, Bunn PA, Jr., Franklin WA, Dziadziuszko R, Thatcher N, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol 2006;24:5034-5042.
52. Hirsch FR, Varella-Garcia M, Cappuzzo F, McCoy J, Bemis L, Xavier AC, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol 2007;18:752-760.
53. Loprevite M, Tiseo M, Chiaramondia M, Capelletti M, Bozzetti C, Bortesi B, et al. Buccal mucosa cells as in vivo model to evaluate gefitinib activity in patients with advanced non small cell lung cancer. Clin Cancer Res 2007;13:6518-6526.
54. Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, et al. K-ras mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res 2007;13:2890-2896.
55. Felip E, Rojo F, Reck M, Heller A, Klughammer B, Sala G, et al. A phase II pharmacodynamic study of erlotinib in patients with advanced non-small cell lung cancer previously treated with platinumbased chemotherapy. Clin Cancer Res 2008;14:3867-3874.
56. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205-216.
57. Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer 1981;47:207-214.
58. Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, et al. PTEN expression and K-ras mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol 2009;27:2622-2629.
59. Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A 2005;102:802-807.
60. Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006;116:2695-2706.
61. Abubaker J, Bavi P, Al-Harbi S, Ibrahim M, Siraj AK, Al-Sanea N, et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene 2008;27:3539-3545.
62. De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of K-ras, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-762.
63. Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 2013;12:220-229.
64. Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther 2012;11:485-491.
Cite this article as: Chen JY, Cheng YN, Han L, Wei F, Yu WW, Zhang XW, Cao S, Yu JP. Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: a systemic review and meta-analysis. Cancer Biol Med 2015;12:126-139. doi: 10.7497/ j.issn.2095-3941.2015.0021
Correspondence to: Jin-Pu Yu
E-mail: Jinpu_yu@hotmail.com
March 30, 2015; accepted June 10, 2015.
available at www.cancerbiomed.org
Copyright ? 2015 by Cancer Biology & Medicine
 Cancer Biology & Medicine2015年2期
Cancer Biology & Medicine2015年2期
- Cancer Biology & Medicine的其它文章
- Paclitaxel-etoposide-carboplatin/cisplatin versus etoposidecarboplatin/cisplatin as first-line treatment for combined small-cell lung cancer: a retrospective analysis of 62 cases
- Current approaches in treatment of triple-negative breast cancer
- Changes in tumor-antigen expression profile as human small-cell lung cancers progress
- Assays for predicting and monitoring responses to lung cancer immunotherapy
- Understanding the function and dysfunction of the immune system in lung cancer: the role of immune checkpoints
- Tumor immune microenvironment characterization and response to anti-PD-1 therapy
